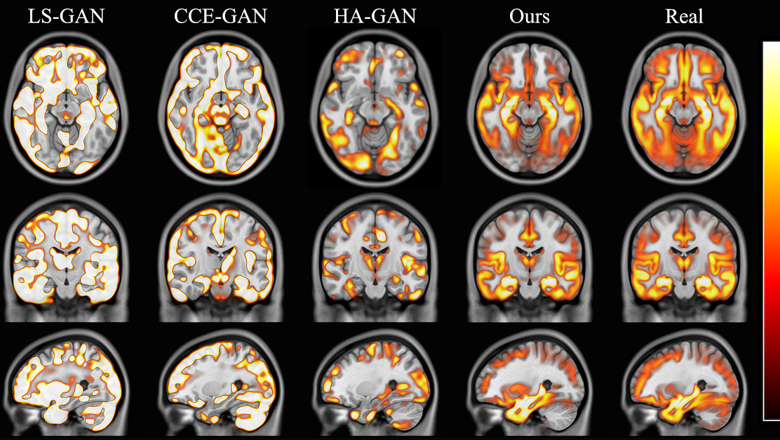
An AI model developed by King's scientists, in close collaboration with University College London, has produced three-dimensional, synthetic images of the human brain that are realistic and accurate enough to use in medical research.
The model and images have helped scientists better understand what the human brain looks like, supporting research to predict, diagnose and treat brain diseases such as dementia, stroke, and multiple sclerosis.
The algorithm was created using the NVIDIA Cambridge-1, the UK's most powerful supercomputer. One of the fastest supercomputers in the world, the Cambridge-1 allowed researchers to train the AI in weeks rather than months and produce images of far higher quality.
The 3D, high-resolution images have all the characteristics of real human brains, such as correct folding patterns and regions of the right size. It can also accurately produce images which reflect clinical factors like age, sex or disease status.
Data produced by the model was realistic enough to replicate human anatomy. For example, the team showed that a dementia research study running on real data would show the same outcomes as a study running on generated synthetic data.
By looking at large volumes of data, the AI model learned how age and sex affect the brain, and how pathologies impact anatomy. These tools have many direct uses, from making AI diagnosis more accurate and equitable, to helping neuroscientists better understand how brains change with age and with disease; this can be transformative to our ability to study the brain and find treatments to critical conditions.
We've taught a computer what the human brain looks like.
Dr Jorge Cardoso, School of Biomedical Engineering & Imaging Sciences
"We've used the algorithm to generate realistic brains for specific ages, sexes and pathologies. Unlike AI images of people with six fingers or three legs, these images are anatomically correct down to every minute detail.
"Collection and access to data from real human brains significantly limits research, and the diversity of real-world data can create research biases and inconsistencies.
"The synthetic images produce high-resolution and potentially infinite data, in which we can control external variables. We can even use the model to make data sets more equitable, such as including a greater range of brains by age, gender or ethnicity.
"The potential for neurological research is enormous. With more development, the technology could help us understand which drugs are best for each patient, how certain conditions might evolve differently in different patients, and how a person's brain might react to a specific treatment.
"Put simply, clinicians would be able to tailor and optimize treatment plans based on the model's predictions for each specific patient."
Parashkev Nachev, Professor of Neurology at UCL said: "To understand the imaged brain we must first acquire the power to recreate it. This implies not merely learning the appearance of a set of brains, but defining the bounds on the possible, counterfactual characteristics of any brain, in health and disease.
"It's a solution which paves the way to overcoming the greatest challenge in medicine: how to predict the optimal treatment for each individual patient to deliver truly personalized care."
With more data and computing power, the models will continue improving. The research is also being expanded to other organs like the heart and lungs, and even to complex multi-system diseases like cancer.
The AI models were developed by King's, UCL and NVIDIA data scientists and engineers, in collaboration with The London Medical Imaging & AI Centre for Value Based Healthcare, with the research funded by UK Research and Innovation, and the Wellcome Innovations-funded Programme for High-dimensional Translation in Neurology.
The research was published in Nature Machine Intelligence.







