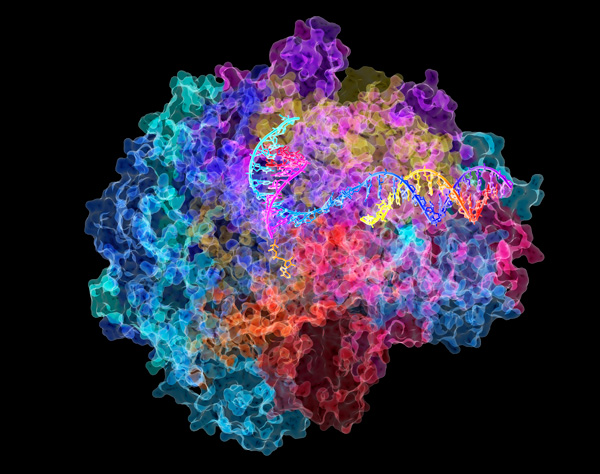
Figure 1: A computer illustration showing the structure of a molecule of RNA polymerase II. Two RIKEN researchers have found an ancient protein fold that clarifies the evolutionary link between ribosomal proteins and RNA polymerase. © DAVID BUSHNELL, KEN WESTOVER AND ROGER KORNBERG, STANFORD UNIVERSITY/SCIENCE PHOTO LIBRARY
In a finding that sheds fresh insights into the early evolution of life on Earth, two RIKEN biologists have conducted lab experiments that have revealed a previously unknown protein fold, which is completely absent in modern proteins1.
The proteins that power essential biological processes such as gene expression and protein production all contain different types of folds known as β-barrel folds. However, the evolutionary pathways between these structures had been unclear.
Now, the discovery in simulations of what is likely a long-lost folding topology-dubbed the double-zeta β-barrel (DZBB)-helps to clarify how complex biomolecular machines might have arisen from simpler precursors.
"The discovery of this missing-link protein fold helps us understand the evolutionary relationship between many different proteins in a much simpler way than we had expected," explains Shunsuke Tagami of the RIKEN Center for Biosystems Dynamics Research (BDR).
DZBB resembles a compact cylinder made up of interlocking protein strands. The ancient, origami-like structure can transform into other key protein shapes with just a few tweaks, Tagami and BDR colleague Sota Yagi found. These DZBB assemblies serve as a versatile foundation for molecular evolution.
Using synthetic biology techniques in the lab, the pair traced the progression of these ancient protein folds. They started with a fold found in DNA and RNA polymerases-enzymes responsible for genome replication and gene transcription. And they showed that, through simple and feasible mutation steps, they might have evolved into the folds found in modern ribosomal proteins, which are essential for synthesizing proteins in cells.
This evolutionary progression required an intermediary structure, DZBB, which could only be uncovered experimentally-it couldn't be predicted through computational methods, even using the latest machine-learning algorithms. This underscores the limitations of current artificial intelligence (AI) models in identifying such complex protein structures.
"Because AI gives answers strongly influenced by the training dataset, experimental validation remains essential to make truly unexpected discoveries," says Tagami.
The results may help solve a long-standing mystery about how primordial proteins evolved to manage genetic processes. DZBB's metamorphic nature, which allows it to adopt multiple stable forms under different conditions, may have allowed molecular machinery in early life to rapidly diversify-much like animal species during the Cambrian explosion.
The findings also raise an intriguing question: if DZBB was so critical to enabling the rise of molecular machines that govern the flow of genetic information within cells, then why is the folding topology no longer seen today?
"DZBB may have been a temporary protein form that existed only during an evolutionary transition between ancient simple forms," says Tagami.
Shunsuke Tagami (left), with colleague Sota Yagi (right), has discovered an ancient protein fold that offers insights into early molecular evolution and protein diversity. © 2024 RIKEN