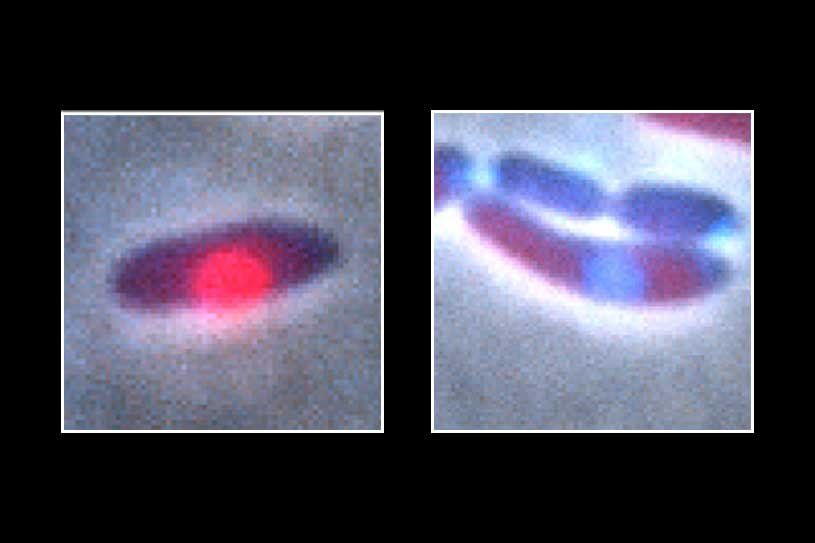
Jumbo phages belong to a group of viruses that attack bacteria. They inject their DNA and then reproduce by taking over the cell's DNA-copying machinery. Eventually, a phage makes so many copies of itself that it will burst bacterial cell it has infected.
Scientists have known for a while that a jumbo phage pulls off its attack and escapes the bacteria's defenses by surrounding its DNA with a protective shield made of protein. But how the shield recognizes certain useful molecules and allows them to pass in while keeping harmful ones out has been a mystery.
Now, research from UC San Francisco, published Feb. 5 in Nature , shows that the protein shield works via a set of "secret handshakes," allowing only a specific set of useful proteins to pass through. The handshakes involve a large, central protein capable of using different parts of itself - like the fingers on a hand - to screen other proteins and grant them passage or not, thereby keeping out the bacteria's defenses.
"This isn't what we expected to see at all," said Joseph Bondy-Denomy , PhD, associate professor of microbiology and immunology at UCSF and senior author of the study. "It's a surprisingly complicated thing for a phage to be doing," he said.
Scientists hope the discovery can help them learn how phages might be used to make new antibiotics that can address the growing crisis of resistance.
Secret handshakes
Jumbo phages belong to a group of viruses called bacteriophages, or phages for short, which were discovered more than a century ago. Initially, phages were seen as a way to treat bacterial infections, because they are harmless to humans and can kill specific bacteria while leaving others alone. Interest died away once antibiotic drugs were developed, but today's urgency to find new ways of fighting antibiotic-resistant bacteria has sparked new research.
Scientists first began working on jumbo phages in the early 1980s but it wasn't until 2017 that researchers at UCSF and UC San Diego worked together to identify the flexible protein that makes up the shield. In 2020, Bondy-Denomy led a study showing that the protein shield protects the phage's DNA from attacks by the bacteria's defenses. He and Claire Kokontis, BS, a graduate student, suspected this shield may give jumbo phages distinct advantages over regular phages when it comes to using these viruses against infections.
The researchers wanted to learn how the shield recognizes bacterial proteins the virus needs to copy its DNA so it can give them passage into the protected area. The secret, they discovered, was a group of proteins made by the phage that interact in an unexpected way. At the center was a phage protein Kokontis called Importer1, or Imp1. For proteins to be imported into the protected space, they had to interact with Imp1.
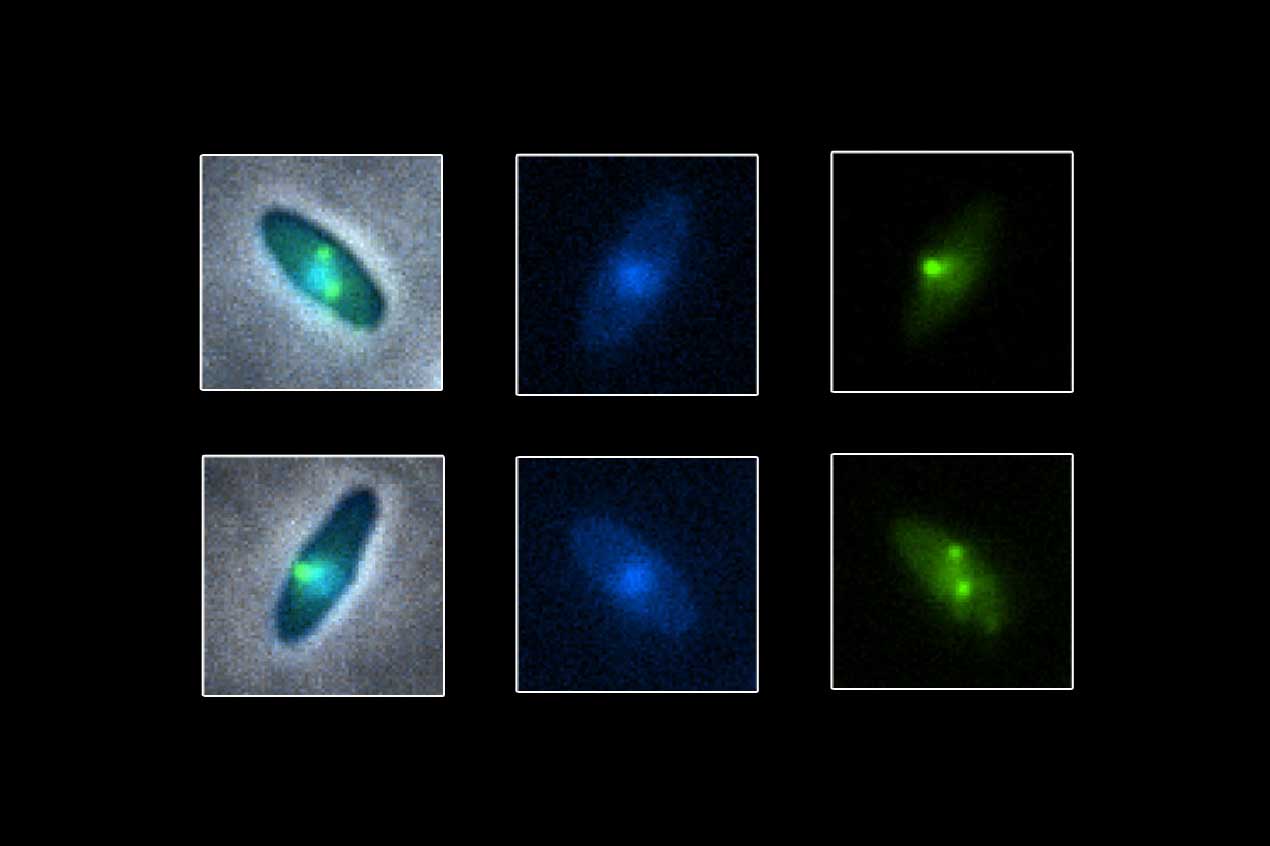
The researchers also found an additional set of importer proteins that assist Imp1 in bringing outside proteins through the shield. The interaction between Imp1 and a protein outside the shield needs to be just right before the protein gets the go-ahead to enter the protected area.
"It's like a secret handshake between two friends," said Bondy-Denomy. "The ones that have the right handshake get the OK, and the others are tossed out."
To see exactly what those handshakes looked like, Kokontis mapped the surface of the Imp1 "hand" at the molecular level. The map revealed that each phage protein that is allowed into the protected area has its own unique way of interacting with the Imp1 hand - one protein touches a thumb, another a finger, another a different finger. This variety of combinations allows the group of importer proteins to recognize an array of handshakes.
A new way of making antibiotics
The researchers did their work using Pseudomonas bacteria, which they chose because it is notorious for its resistance to most antibiotics.
What they learned will help scientists improve on an old approach that was left behind once antibiotics became standard. Called phage therapy, it involves fighting one infection with another. First a human gets infected by bacteria. Then the human uses a phage to infect and kill the bacteria.
But bacteria are quick to evolve new defenses. Once they have devised a way to get past the phage's protective shield, they will kill the phages. Understanding exactly how the shield's secret handshakes work will help scientists engineer phages that can withstand these evolutionary changes.
Bondy-Denomy's lab has already developed a CRISPR-based, gene editing method to make the necessary genetic changes to these jumbo phages. Scientists can also employ that knowledge and ability to engineer jumbo phages that produce drugs or fight cancers caused by bacterial infections.
"We're just at the starting point of realizing all this potential," Kokontis said. "By getting a handle on the basic science of how these phages work, we're laying the groundwork to adapt them for fighting disease."
Authors: Other authors of this study are Timothy Klein and Sukrit Silas of UCSF
Funding: This work was funded by the NIH (grants R01 AI171041 and R01 AI167412).






