Nanozymes have high catalytic activity, high stability and high adaptability, and have become a new sensitive material for building sensors in the field of detection. Designing and preparing efficient nanozymes and promotion of their application in food detection have attracted much attention from researchers.
Recently, a team led by Prof. HUANG Qing from the Hefei Institutes of Physical Science of the Chinese Academy of Sciences, prepared a series of Cu metal organic framework (MOF) nanozymes with the gas-liquid interface dielectric barrier discharge (DBD) low-temperature plasma (LTP) technology.
These nanozymes have different base ligands and mimic the activity of laccase. They exhibited varying responses to five common bioactive substances found in food.
HUANG and his team also developed encoded array sensors capable of high-throughput, sensitive, and rapid identification and quantitative analysis of substances in the concentration range of 1.5-150 μg/mL. The sensors will be used for intelligent sensing and identification of bioactive components in food, according to the researchers.
The degree of color change induced by the nanozymes can be easily observed using a smartphone, with these sensors, enabling portable and intelligent rapid food detection.
This study provides not only a new way to prepare efficient nanozymes, but also an intelligent and convenient method for food inspection.
The relevant research results were published in Biosensors and Bioelectronics.
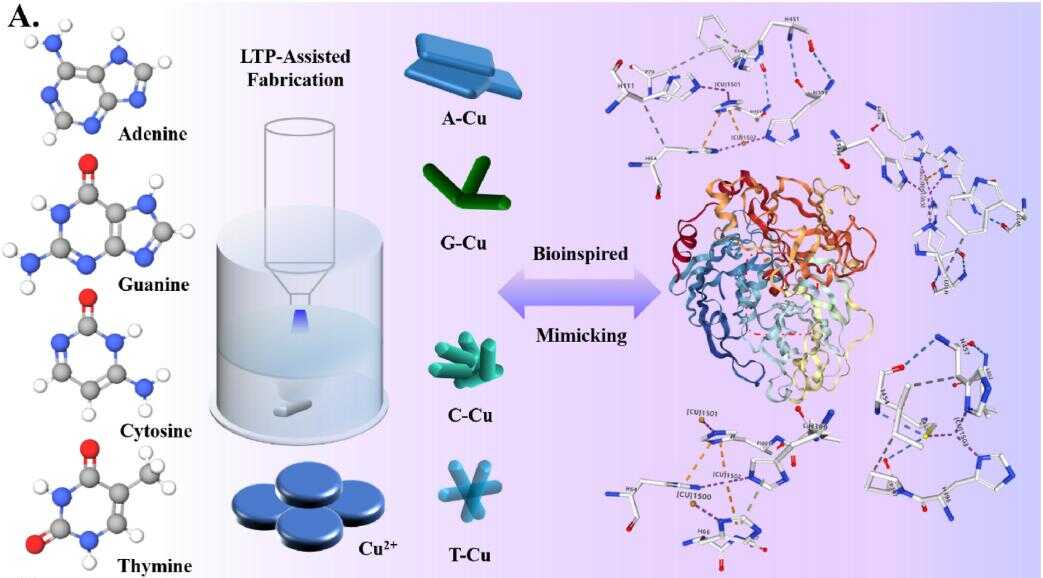
Schematic diagram of Cu-MOF nanozyme design and LTP preparation process. (Image by LIU Chao)
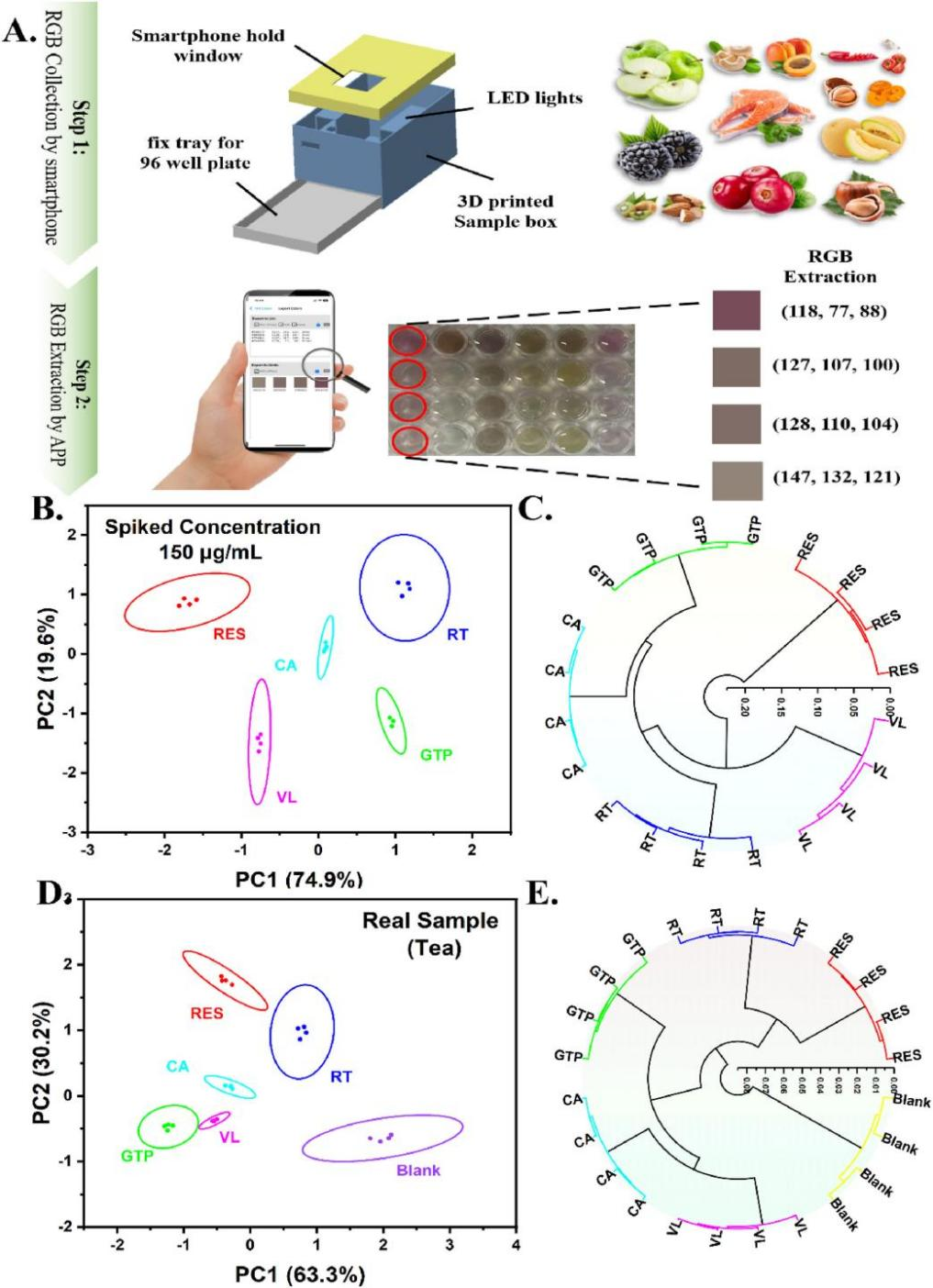
Evaluation of natural active ingredients in real food samples using smartphones. (Image by LIU Chao)






