The genetic code of all three kingdoms of life is universal and encodes the same 20 natural amino acids for a variety of complex physiological functions. The expansion of the genetic code by incorporating UAAs with diverse functional groups has enabled the synthesis of proteins with enhanced or novel functions, and the construction of UAA-dependent synthetic auxotrophs. However, it is extremely challenging have an in vivo site-specific UAA incorporation system with comparable efficiency and similar protein yield when UAAs are used in place of natural amino acids. In 2020, LIN Shixian et al. reported the design of broadly orthogonal chimeric translation system that functions in bacterial and mammalian systems for genetic code expansion of post-translational modification and fluorescent UAAs. However, like all current orthogonal translation systems, the efficiency of chimera system is much lower than endogenous translation systems for natural amino acids, thereby inhibiting its application in protein design and functional studies.
On December 2, the team led by Prof. LIN Shixian at the Zhejiang University Life Sciences Institute published an article entitled "Directed-evolution of translation system for efficient unnatural amino acids incorporation and generalizable synthetic auxotroph construction" in the journal Nature Communications. In this paper, the researchers reported the directed-evolution of translation systems that allowed the incorporation of unnatural amino acids with similar efficiency to natural amino acids and the construction of synthetic auxotroph in a generalizable way.
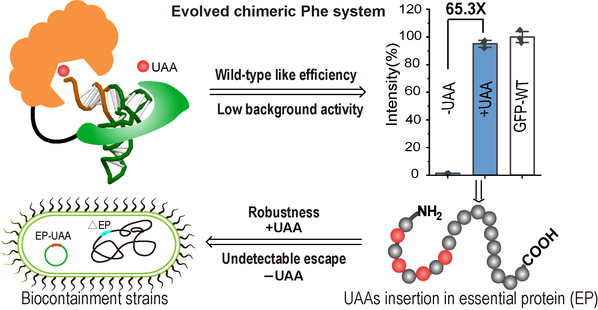
Efficient unnatural amino acids incorporation and generalizable synthetic auxotroph construction with the evolved chimeric Phe system
In this study, Lin and his team engineered a chimeric phenylalanine (Phe) system with improved incorporation efficiency that achieved wild-type like efficiency in the production of full-length proteins and had low background activity in the absence of UAAs. These desirable characteristics were demonstrated with multiple Phe and tryptophan (Trp) analogs by directed-evolution of the chimeric Phe tRNA (chPheT) at the acceptor stem and the chimeric Phe synthetase (chPheRS). After screening a library of ~1.7 × 107 tRNA molecules and 13,000 synthetase clones, the researchers obtained mutants that could significantly enhance the efficiency of the chimeric translation system in recognizing UAAs. The directionally evolved chimeric translation system was able to encode multiple UAAs at many sites in the model protein and in multiple functional proteins almost as efficiently as that of natural amino acids. Meanwhile, it exhibited remarkably high fidelity. Impressively, the engineered chPheRS/ chPheT pairs showed a 65-fold increase in amber suppression efficiency. The engineered chPheRS/chPheT pair for AzF incorporation was then introduced into a commercial E. coli strain to construct an UAA-dependent synthetic auxotroph. "By introducing in-frame amber codons into the essential genes, we were able to identify several synthetic auxotroph strains that were strictly dependent on exogenously supplied UAAs for growth," said Lin.
"This study provides a powerful and general technology for the synthesis of UAA-dependent synthetic auxotrophs for the containment of genetically modified organisms and the development of live-attenuated vaccines," Lin added.






