With a novel approach, EMBL scientists discovered important interactions between molecular machines, potentially offering new opportunities for drug development
Summary
- A new study by EMBL's Duss Group sheds light on the dynamic interactions between two essential cellular processes - transcription and translation - in bacteria.
- The researchers created real-time movies capturing important interactions between the key molecular components of these processes - RNA, RNA polymerases, and ribosomes.
- Understanding how these fundamental cellular mechanisms work in bacteria offers new ways to think about drug development - especially in the context of growing antimicrobial resistance.
Choosing a film for a movie night is always a battle. Now imagine if you could pick one that provided a window into some of the most fundamental biological processes that keep us alive. For the first time ever, researchers have captured a real-time molecular movie to show how two essential cellular processes - transcription and translation - interact with each other in bacteria.
In all living organisms, DNA contains the code that defines cellular structures and functions. An enzyme called RNA polymerase deciphers this code and converts it into RNA, a molecule that closely resembles DNA. This transfer of life's code from DNA to RNA is called transcription. Next, a molecular machine called 'ribosome' uses the message encoded in RNA to build proteins - the molecules performing most of the essential functions of our cells. This process is called translation.
"In bacterial cells, transcription and translation take place in the same cellular compartment," explained Olivier Duss, Group Leader at EMBL Heidelberg and senior author of the new study. "In human cells, transcription is localised to the nucleus - the compartment where DNA is stored, separated from the rest of the cell by a membrane. The transcribed RNA is then transported outside the nucleus to be translated into proteins, which exclusively happens in the cytoplasm - the cellular compartment surrounding the nucleus. Bacterial cells are much simpler in their cellular structures, and lack a nucleus, thus allowing transcription and translation to happen not only in the same place but also at the same time."
Scientists have previously characterised transcription and translation as single processes, but how the two interact is not well understood. This was partially because such studies relied on techniques like cryo-electron microscopy, which require frozen samples, thus only providing snapshots of the process.
This problem interested the Duss Group , which uses single-molecule technologies, structural biology, and biochemistry to understand how large molecular machines involved in crucial cellular functions cooperate with each other.
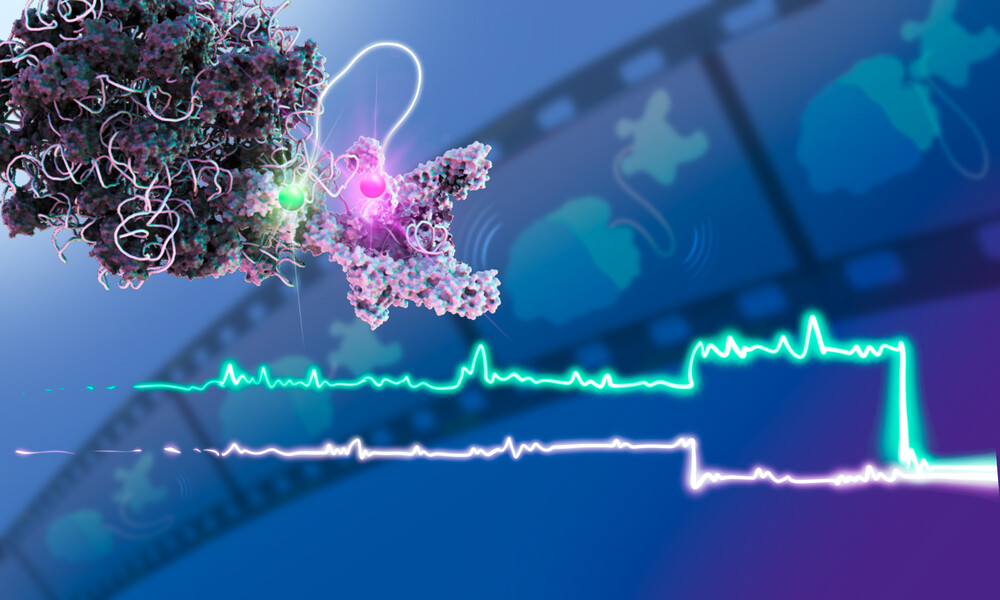
To investigate how transcription and translation work together, the research team, co-led by research scientist Nusrat Qureshi, artificially recreated the cellular environment required for these processes to take place. This allowed them to closely track the dynamics of interacting RNA polymerases and ribosomes, one pair at a time, using a technique called single-molecule multi-colour fluorescence microscopy.
Simply put, the technique works by tagging the RNA polymerase and the ribosome with small chemicals that act as proximity sensors. When the two molecules interact, they emit a signal that can be captured by a fluorescent microscope. When they stop interacting, the signal disappears.
Using this, the scientists captured several minutes of the dynamic interplay between RNA polymerase and the ribosome. For the first time ever, they could look through a microscope and simultaneously watch transcription and translation in action.
"I'm very excited that we can finally watch the entire process," Duss said. "We can put these snapshots into motion, and that lets us better understand how the two machineries cooperate. By putting it all together, we start seeing emerging behaviours that cannot be predicted otherwise."
One such emergent behaviour the scientists discovered was that the RNA polymerase and the ribosome can communicate even at a distance, with a rather long stretch of looping RNA connecting them.
In this, the two molecular machines act much like a pair of mountain climbers tethered by a long rope. The rope is loose enough to prevent collisions with one another but tight enough to let each climber help the other when needed.
The team also observed that transcription is more efficient when translation occurs at the same time. In other words, when an active RNA polymerase is followed on the same RNA molecule by a progressing ribosome, its productivity is higher.
"It is beautiful to be able to watch how these processes work together. Any person working in a team knows the importance of collaboration," Duss said. "If everyone tries to just work on their own, their efficiency will be much lower. It seems like the cell's molecular machines know this too."
The new study was published today in the journal Nature .
While this study focused on isolated molecules in an artificial set-up, the Duss Group is now preparing to expand their understanding of this process to live cells. As part of their recently awarded ERC Consolidator Grant , they also plan to include additional cellular processes in the study to see if the 'climbing' coordination involves more than just two partners.
Shedding light on how fundamental cellular mechanisms work in bacteria paves the way to developing new ways to fight bacterial pathogens at a time when antibiotic resistance is an important health issue. Researchers can potentially go beyond standard antibiotics, preventing resistance issues by cooperatively targeting two cellular machines rather than just one.
"This work is a great example of the importance of basic research in the broader context," Duss said. "Basic research is what helps us understand how biology works, which then translates into new discoveries like novel drugs, advanced treatments, and better opportunities."






