Medaka fish that lacked functional Hmgn2 genes were unable to distinguish between simple shapes, revealing a new function for the regulatory gene.
Shape preferences of wild type medaka (WT) vs. oHmgn2 knockout mutants (ohmgn2 mutant) in a Y maze with triangles in one arm (blue) and circles in the other (pink). The WT medaka exhibits a preference for triangles over circles, spending more time in that arm of the maze, while the ohmgn2 mutant spends an almost equal time in both arms of the maze. Video plays at 8X speed. (Provided by Saori Yokoi)
The medaka, also known as the Japanese rice fish (Oryzias latipes), is a model organism for the study of biology, as well as a popular aquarium fish. As a model organism, much research has been carried out to understand all aspects of the medaka, but much still remains to be done, especially in the area of genetics.
A research team led by Assistant Professor Saori Yokoi of the Faculty of Pharmaceutical Sciences, Hokkaido University, has discovered an evolutionarily distinct variant of the gene Hmgn2 in medaka, which influences shape preference in the species. Their findings were published in the journal Communications Biology.
"We identified this Hmgn2 variant, oHmgn2, as part of our research on identifying genes expressed in the medaka brain with unknown functions," Yokoi explains. "HMGN proteins play crucial roles in chromatin remodeling and gene expression regulation in other vertebrates."
oHgmn2 was selected as it was an unknown gene that was characteristically expressed in medaka brains. After confirming that the gene coded for an HMGN protein, sequencing oHmgn2 RNA showed that it was 99 base pairs shorter than Hmgn2 from other vertebrates. Evolutionary analysis confirmed that oHmgn2 was a unique variant.
"The most striking difference was the presence of oHmgn2 in the nucleolus, the region in the nucleus where transcription from DNA to RNA occurs," Yokoi elaborates. "oHmgn2 is predicted to have a weaker interaction with DNA-binding proteins called histones, compared to the typical interaction, explaining its distribution."
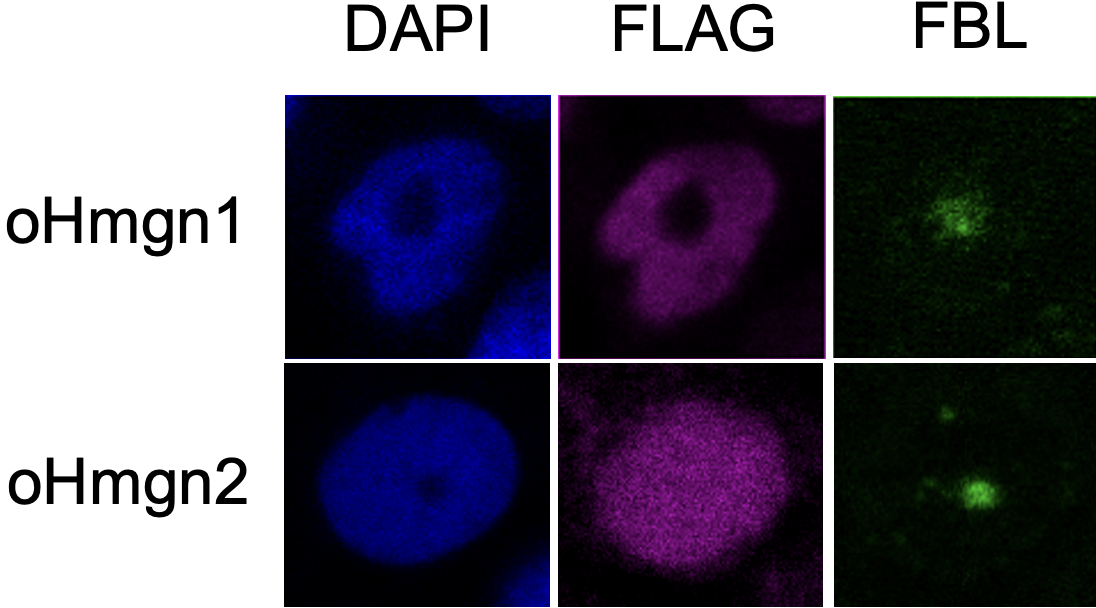
oHmgn2 shows different nuclear localization compared to related Hmgn proteins (FLAG, purple). oHmgn1 is associated with the nucleoplasm (DAPI, blue) of the nucleus, and is not associated with the nucleolus (FBL, green). However, oHmgn2 is dispersed throughout the nucleus. (Shuntaro Inoue, Yume Masaki, Shinichi Nakagawa, Saori Yokoi. Communications Biology. August 23, 2024)
oHmgn2 expression is localized in the neurogenic regions of the brain, and the team found indications that it was involved in neural progenitor cell function. Knockout studies showed that the lack of functional oHmgn2 affected the development of the telencephalon-the region of the brain that corresponds to the cerebrum/frontal lobes in humans.
Most interestingly, wild type medaka exhibits a preference for triangles over circles, spending more time in that arm of the maze, while oHmgn2 knockout medaka spend an almost equal time in both arms of the maze-indicating deficits in their ability to recognize shapes.
"Our study has revealed that the oHmgn2 gene is a key player in the molecular evolution of brain development and cognitive behavior within the medaka," Yokoi concludes. "Future work will focus on understanding the mechanisms by which oHmgn2 affects shape recognition in medaka."

The research team, including Saori Yokoi (standing, second from left), Yume Masaki (standing, right) and Shuntaro Inoue (seated, right). (Photo: RNA Biology Laboratory, Faculty of Pharmaceutical Sciences, Hokkaido University)
Original Article:
Shuntaro Inoue, Yume Masaki, Shinichi Nakagawa, Saori Yokoi. An evolutionarily distinct Hmgn2 variant influences shape recognition in Medaka Fish. Communications Biology. August 23, 2024.
DOI: 10.1038/s42003-024-06667-8
Funding:
This work was supported by the National Institute for Basic Biology (NIBB) Collaborative Research Program (23NIBB336); the Japan Society for the Promotion of Science (JSPS) KAKENHI (21H05708, 23K05841, 23H03839); Astellas Foundation for Research on Metabolic Disorders; Takeda Science Foundation; and, the Naito Foundation.







