Researchers at Kanazawa University report in the Journal of Physical Chemistry Letters high-speed atomic force microscopy studies that shed light on the possible role of the open reading frame 6 (ORF6) protein COVID19 symptoms.
————————————————————————————————————————————————————————————————
While many countries across the world are experiencing a reprieve from the intense spread of SARS-CoV-2 infections that led to tragic levels of sickness and multiple national lockdowns at the start of the decade, cases of infection persist. A better understanding of the mechanisms that sustain the virus in the body could help find more effective treatments against sickness caused by the disease, as well as arming against future outbreaks of similar infections. With this in mind there has been a lot of interest in the accessory proteins that the virus produces to help it thrive in the body.
"Similar to other viruses, SARS-CoV-2 expresses an array of accessory proteins to re-program the host environment to favor its replication and survival," explain Richard Wong at Kanazawa University and Noritaka Nishida at Chiba University and their colleagues in this latest report. Among those accessory proteins is ORF6. Previous studies have suggested that ORF6 potently interferes with the function of interferon 1 (IFN-I), a particular type of small protein used in the immune system, which may explain the instances of asymptomatic infection with SARS-CoV2. There is also evidence that ORF6 causes the retention of certain proteins in the cytoplasm while disrupting mRNA transport from the cell, which may be means for inhibiting IFN-I signalling. However, the mechanism for this protein retention and transport disruption was not clear.
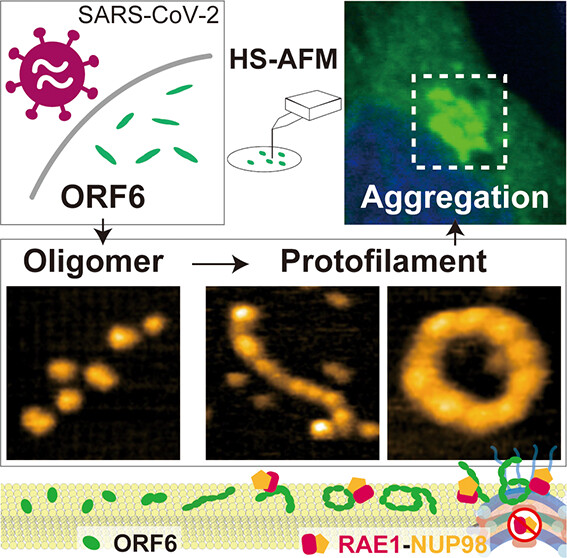
To shed light on these mechanisms the researchers first looked into what clues various software programs might give as to the structure of ORF6. These indicated the likely presence of several intrinsically disordered regions. Nuclear magnetic resonance measurements also confirmed the presence of a very flexible disordered segment. Although the machine learning algorithm AlphaFold2 has proved very useful for determining how proteins fold, the presence of these intrinsically disordered regions limits its use for establishing the structure of ORF6 so the researchers used high-speed atomic force microscopy (HS-AFM), which is able to identify structures by "feeling" the topography of samples like a record player needle feels the grooves in vinyl.
Using HS-AFM the researchers established that ORF 6 is primarily in the form of ellipsoidal filaments of oligomers - strings of repeating molecular units but shorter than polymers. The length and circumference of these filaments was greatest at 37 °C and least at 4 °C, so the presence of fever could be beneficial for producing larger filaments. Substrates made of lipids - fatty compounds – also encouraged the formation of larger oligomers. Because HS-AFM captures images so quickly it was possible to grasp not just the structures but also some of the dynamics of the ORF6 behavior, including circular motion, protein assembly and flipping. In addition, further computer analysis also revealed that the filaments were prone to aggregate into amyloids as found in some neurodegenerative diseases, and which can lead to complications in COVID19 symptoms. As the researchers point out this aggregation works "to effectively sequester a vast numbers of host proteins, particularly transcription factors involved in IFN-I signaling."
Since these filaments break up in the presence of certain alcohols, urea, or sodium dodecyl sulfate Wong, Nishida and their colleagues conclude that the protein is largely held together by hydrophobic interactions. "Potential druggable candidates that dissociate ORF6 aggregates by disrupting hydrophobic interactions should be considered and tested in the near future to evaluate their therapeutic value in COVID19 management and treatment."
Figure: Observation of aggregation dynamics by COVID-19 severity factor ORF6 protein.
©Nishide, et al., 2023 Journal of Physical Chemistry Letters
Glossary
High-speed atomic force microscopy
Atomic force microscopy (AFM) "feels" the topography of surfaces by the increase and decrease of surface forces tugging on a nanoscale tip as changes in surface height change the distance between the tip and the surface. The tip is mounted on a cantilever so that the tiny changes in the force can be read out by the resulting deflection in the cantilever.
AFM can resolve structures with subnanometre scale resolution. It has particular advantages for biological studies because it does not require conducting substrates or a current, which are requirements for other primary microscopes with comparable resolutions, such as the scanning tunnelling microscope.
For a long time AFM was limited by the time it takes to capture an image of the surface, but this changed when Toshio Ando at Kanazawa University revealed how the tool could be upgraded to high-speed AFM using various modifications to the scanning, deflection detection and other electronic devices, as well as specifications for the cantilever. With high-speed AFM it became possible to capture dynamics at the nanoscale for the first time.
Reference
Authors: Goro Nishide, Keesiang Lim, Maiki Tamura, Akiko Kobayashi, Qingci Zhao, Masaharu Hazawa, Toshio Ando, Noritaka Nishida, Richard W. Wong.
Title: Nanoscopic Elucidation of Spontaneous Self-Assembly of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Open Reading Frame 6 (ORF6) Protein
Journal: Journal of Physical Chemistry Letters
Published on Sep. 14,2023
DOI: 10.1021/acs.jpclett.3c01440
URL: https://pubs.acs.org/doi/10.1021/acs.jpclett.3c01440