How soft or rigid proteins are determines how quickly they enter the cell nucleus, according to new research from King's College London and the Francis Crick Institute.
The findings open new ways to enhance the transport of molecular cargos into the cell nucleus, which could help gene control.
Proteins enter in and out of the nucleus-the control centre of cells-by crossing a large and highly selective molecular channel called the nuclear pore complex. Molecular traffic through the nuclear pore complex is critical for many cellular functions, ranging from instructing cells to divide to telling the nucleus to switch on or off certain genes.
The paper, published today in Nature Physics, shows that the mechanical properties -how easily they can be mechanically unraveled- of the transported proteins can dictate how quickly they enter the nucleus.
"We've made a fundamental discovery that the mechanics of a protein - how soft or how much it deforms, dictates the way it enters a cell's nucleus, and therefore how quickly and effectively it instructs the nucleus to carry out a function."
Professor Sergi Garcia-Manyes, Professor of Biophysics
Led by King's Professor of Biophysics, Professor Sergi Garcia-Manyes, the findings complement previous research in the field, which showed that the size and composition of proteins can change how easily they cross to the nucleus to carry out their function.
Professor Sergi Garcia-Manyes said: "We've made a fundamental discovery that the mechanics of a protein - how soft or how much it deforms, dictates the way it enters a cell's nucleus, and therefore how quickly and effectively it instructs the nucleus to carry out a function.
"Although we've only looked at the nuclear pore, this mechanism could regulate entry into other parts of the cell, such as the mitochondria or proteasomes. Knowing that a more flexible protein can enter the nucleus quicker could even inform the development of new biotechnological tools for pharmaceuticals."
The study involved tracking the movement of proteins in single cells. For proteins that were the same size and had the same amino acid composition, the speed at which they could cross was influenced by the mechanical stability near the protein's 'nuclear-localisation sequence', which is the chemical tag that signals the protein to the nucleus.
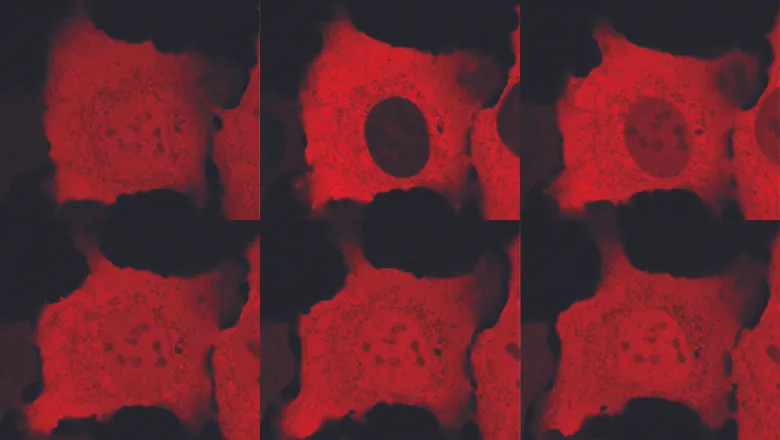
Video Caption: Timelapse of an experiment called an optogenetic experiment, which allowed the researchers to track protein movement in cells. Light is shone on the cells and the red fluorescent protein leaves the nucleus. The light is then switched off and the red fluorescent protein re-enters the nucleus over time. The video takes place over 55 minutes. Credit: Natalie Milmoe, Single Molecule Mechanobiology Laboratory, the Francis Crick Institute.
Once it was identified that proteins with a soft or flexible region next to this sequence were able to cross into the nucleus more quickly, the team engineered a soft tag that could be added near the sequence on stiffer proteins to help them enter more easily.
This was tested by tagging a transcription factor - a protein that regulates the expression of certain genes to dictate specific cellular functions - called MRTF-A, which controls cellular motility.
Lecturer in Biological Physics Dr Rafael Tapia-Rojo and co-first author of the paper said: "Our findings were rather unanticipated, and it was striking to see how measurements at the single molecule level can be so directly linked to what happens at a cellular level, using a newly designed optomechanical approach."
The researchers are now investigating how transcription factors have evolved to contain flexible regions that allow them to enter the nucleus more easily.








