In a recent study published in "Nature Communications", the Vorholt lab investigated the properties of plant microbiota involved in host protection against pathogen colonization. They identified the presence of specific strains that confer robust protection across different biotic contexts.
Plant-associated microbiomes contribute to important ecosystem functions such as host resistance to biotic and abiotic stresses. The factors that determine such community outcomes are inherently difficult to identify under complex environmental conditions. In their study, the research team of the Vorholt lab introduces an experimental and analytical approach to explore microbiota properties relevant for a microbiota-conferred host phenotype, here plant protection, in a reductionist system. They examined randomly assembled synthetic communities (SynComs) of five bacterial strains each, and then performed classification and regression analyses as well as empirical validation to test potential explanatory factors of community structure and composition, including evenness, total commensal colonization, phylogenetic diversity, and strain identity.
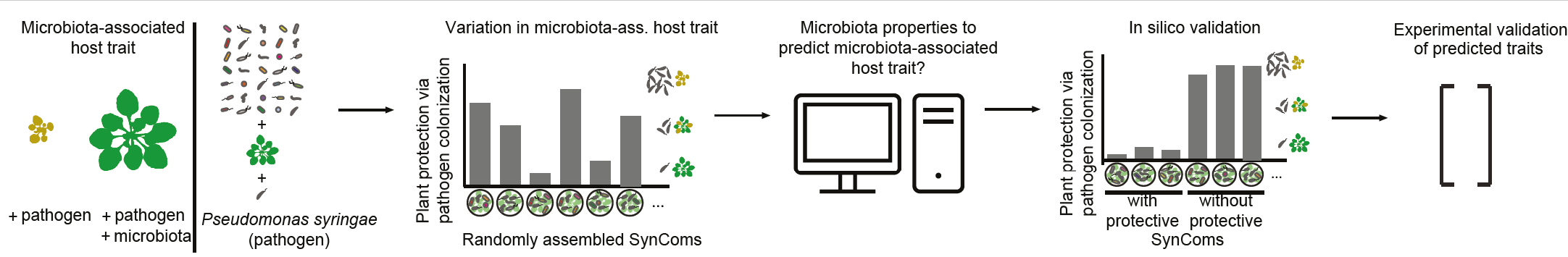
The authors found that strain identity was an important predictor of pathogen reduction, with machine learning algorithms improving performance compared to random classifications and non-modelled predictions. Experimental validation confirmed the identified strains as the main drivers of pathogen reduction and additional ones that conferred protection in combination. Beyond the specific application presented in their study, the work provides a framework that can be adapted to help determine features relevant to microbiota function in other biological systems.
Link to the paper in external page"Nature Communications".






