Much like humans generate mountains of garbage, our cells are constantly discarding proteins that are damaged or no longer needed. The cellular waste disposal system called the proteasome is best known for its central role in protein degradation and recycling, but as far back as the 1990s it was shown that products of this process - short protein sequences called peptides - can be displayed on the cell surfaces, helping the immune system to identify threats. In a new study published today in Nature , Prof. Yifat Merbl 's lab at the Weizmann Institute of Science reports uncovering a surprising immune mechanism involving the proteasome. The team discovered that some of the peptides released in the proteasome during protein breakdown are capable of killing bacteria. These findings expand our understanding of the body's innate defenses and offer new hope for tackling the growing threat of antibiotic resistance.

Several years ago, scientists in Merbl's lab in Weizmann's Systems Immunology Department developed an innovative technology that enabled them to "dumpster dive" within the proteasome, a complex molecular machine composed of numerous proteins. Using this advanced tool, the researchers tracked proteasomes under various disease conditions, such as lupus and cancer, accumulating vast amounts of data on the degraded protein fragments.
"We took a broad look at all the data and asked ourselves: Could the products of the degradation play an additional role, beyond being presented to the immune system?" says Merbl of the new study's starting point. To their surprise, the researchers found that many of these degradation products matched sequences previously identified as antimicrobial peptides, critical components of the innate immune system, which acts as the body's first line of defense against bacteria, viruses and parasites. For years, it was known that such peptides may be generated by protein-cutting enzymes called proteases that "released" them from proteins so they could become active, but the new findings of Merbl's lab have shown that such peptides may be activated by proteasomes. In fact, the study revealed that the proteasome itself constantly produces these peptides as part of its routine activity, and that this production ramps up significantly during bacterial infections.
""This peptide database opens a new frontier for developing personalized treatments against infections and other medical conditions"
"Before now, we knew nothing about the connection between proteasome products and the production of these peptides," says Merbl. "In light of our findings, we conducted an extensive series of experiments demonstrating that the proteasomes are key to this defense system." In one experiment conducted on human cells, the researchers inhibited the proteasomes in one group of cells and left them untouched in the other group; when the cells were infected with salmonella, the invading bacteria thrived in the group that lacked active proteasomes. In another experiment, bacteria thrived when the proteasome functioned normally but the peptides produced within it were destroyed.
The effectiveness of the peptides was also demonstrated in mice infected with bacteria that cause pneumonia and sepsis, a life-threatening condition triggered by an immune response to severe infection. Experiments in these mice showed that treatment with a proteasome-derived peptide significantly reduced the number of bacteria, lessened tissue damage and even improved survival rates. The results surprised the researchers for two reasons. First, they showed that a single peptide that is naturally made by the body can prove effective against a life-threatening condition when administered in large amounts. Second, the results of the treatment were comparable to those of treatment with strong antibiotics in clinical use.
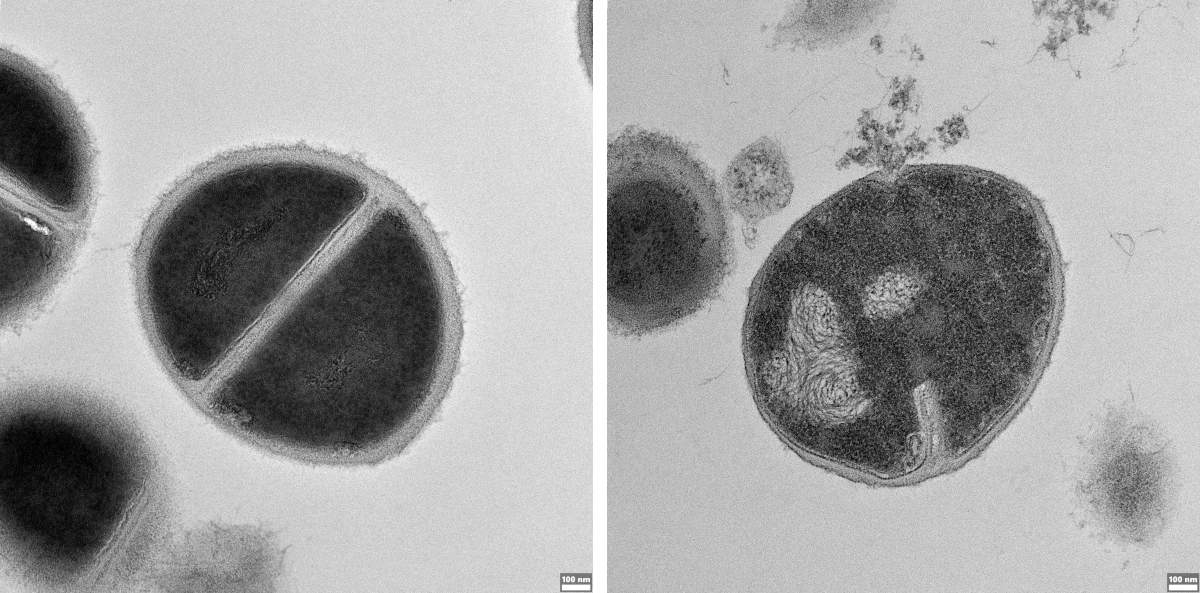
The researchers were most excited, however, when they realized that bacterial infection sends the proteasome into "turbo mode." "We saw that infection causes the proteasome to change its protein-cutting mode, 'favoring' the production of peptides with antibacterial properties," Merbl says. When the researchers tried to explain what caused this change, within an hour of infection they identified proteasomes with a control unit called PSME3 and found that this subunit was responsible for prioritizing the production of such peptides. When they prevented the proteasomes from using this control subunit, the bacteria were less damaged, which highlights the importance of the proteasome in providing a first line of defense against infection.
"The ability to track how the activity of the proteasome changes in response to bacterial infection was based on the technology we developed several years ago," explains PhD student Karin Goldberg, who led the project. "The turning point came when we saw that the proteasome's peptide-cutting activity changed during infection. That was when we realized we had uncovered a previously unknown immune mechanism."
The researchers then asked a broader question: How many hidden antimicrobial peptides might be lurking within human proteins? Using an algorithm to analyze all the proteins made by the human body, they identified peptides with potential antibacterial properties in 92 percent of human proteins. Their simulations revealed over 270,000 previously unknown peptides that could be released by the proteasome, representing a huge untapped reservoir of natural antimicrobial agents.
"This peptide database opens a new frontier for developing personalized treatments against infections and other medical conditions," Merbl explains. For instance, natural peptides could be tailored to strengthen immune defenses in patients with weakened immunity, such as organ transplant recipients or cancer patients. Moreover, as antibiotic resistance continues to pose a major public health challenge, the study's findings not only redefine our understanding of cellular immunity but also pave the way for innovative therapies based on natural mechanisms.
Science Numbers
Cellular trash bins - protein complexes known as proteasomes - make up between 1 and 2 percent of the protein content in a cell. They are responsible for degrading 70 percent of the cellular proteins.
Beyond the clinical implications, Merbl says that the greatest thrill was discovering a fundamental cellular mechanism that is regulated by the proteasome and is different from anything previously known: "This study highlights how technological innovation and basic research intertwine in unforeseen ways. Without the technology that allowed us to analyze the cellular trash, we would not have made this discovery, but when we developed this technology, we never imagined that we would uncover a new immune mechanism."
Also participating in the study were the teams of Prof. Zvi Hayouka from the Hebrew University of Jerusalem, Prof. Nissan Yissachar from Bar-Ilan University and Prof. Gee W. Lau from the University of Illinois.
Prof. Merbl's research is supported by the Dr. Barry Sherman Institute for Medicinal Chemistry; the Moross Integrated Cancer Center; the EKARD Institute for Cancer Diagnosis Research; and the Dr. Gilbert S. Omenn and Martha A. Darling Weizmann Institute - Schneider Hospital Fund for Clinical Breakthroughs through Scientific Collaborations.






