Yulan Xiong and her team have discovered a much-searched-for regulator that plays a key role in genetic causes of Parkinson's Disease

Image by Peggychoucair from Pixabay.
Yulan Xiong, associate professor of neuroscience at UConn Health, and her team have discovered one more piece of the puzzle of the genetic causes of Parkinson's Disease, paving the way for new treatment options.
A mutation on a gene called LRRK2 is the most common genetic cause of Parkinson's Disease, which affects nearly one million Americans.
While scientists have known that LRRK2 mutations are important to understanding Parkinson's Disease for years, the mechanism of how mutations cause disease development is still poorly understood.
Xiong and her team previously discovered that an enzyme called ATIC and its substrate (AICAR) regulates LRRK2 on the mRNA level during the process in which DNA is transcribed into RNA and then expressed as a protein. ATIC is overactive in patients with Parkinson's Disease and instructs LRRK2 to make too much of a protein called daradarin.
Building on that work, Xiong has now discovered a key regulator that could be used to inhibit the overactivity of LRRK2. Xiong published these findings in Science Advances.
LRRK2 has two enzyme "domains" - a kinase and GTPase domain. Kinase is responsible for catalyzing the transfer of phosphate groups. GTPase binds to the nucleotide guanosine triphosphate (GTP) and guanosine diphosphate (GDP) in the protein regulation process. Xiong has identified a key regulator for GTPase function called CalDAG-GEFI (CDGI) to control the switch of binding GTP or GDP of LRRK2 GTPase.
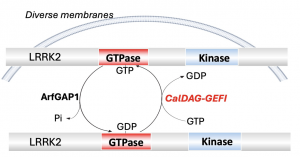
Most other research has focused on understanding and targeting the kinase domain because it is an easier pharmaceutical target than GTPase. While LRRK2 GTPase is a crucial target for Parkinson's Disease research, it is difficult to selectively modulate GTPases with drugs. Further, one part of the GTPase domain, the COR domain, has no known small molecule binding sites where drugs could attach themselves.
"This is a very understudied area," Xiong says. "There is not much work on this."
This is a major discovery, as researchers have spent years searching for this missing regulator.
"The significance is that we identified this key regulator that can, basically, switch on or switch off LRRK2's GTPase function," Xiong says.
If they can inhibit this regulator, scientists can stop the overactivity of daradin to slow the progression of Parkinson's Disease.
Xiong completed this research using cell and mouse models. The next step will be completing studies using human samples.
Xiong has also been collaborating with an external company to develop a small molecule capable of passing the blood brain barrier (BBB) to deliver potential drugs based on this work.
They are also working to make the inhibitor more targeted to regulate just the activity of LRRK2.
"The inhibitor we identified previously targets LRRK2 protein expression," Xiong says.
Qiufang Liu at UConn Health is the first author of this work. This study collaborated with Noah Guy Lewis Guiberson, Ted M. Dawson, and Valina L. Dawson at Johns Hopkins; Jill R. Crittenden and Ann M. Graybiel at MIT; and Gang Ma and Jianzhong Yu at UConn Storrs. Other researchers who contributed to this study are Bingxu Huang, Shifan Chen, Dong Zhu, and Xin-Ming Ma at UConn Health.
This work was funded by the National Institutes of Health, National Science Foundation, the Parkinson's Foundation, William N. & Bernice E. Bumpus Foundation, JPB Foundation, and the UConn Health Startup Fund.






