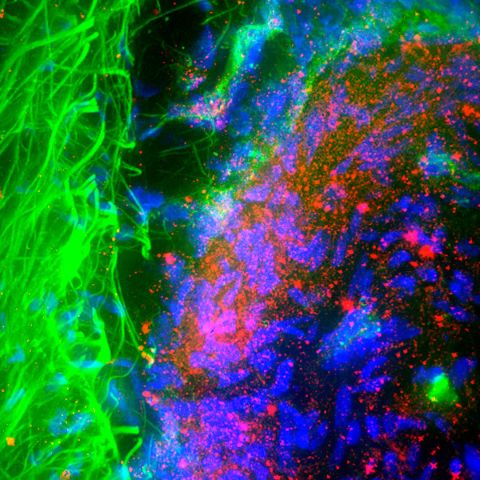
Figure 1: A fluorescent micrograph of a section of brain affected by Parkinson's disease. Neuron nuclei are blue, while the protein alpha-synuclein is red. A team that included a RIKEN researcher has enhanced the accuracy of a lab test for measuring alpha-synuclein levels. © MYA C. SCHIESS, ROGER BICK, UT MEDICAL SCHOOL/SCIENCE PHOTO LIBRARY
A team that includes a RIKEN researcher has refined a lab test for measuring protein aggregate levels in samples from patients with certain neurodegenerative diseases1. This has the potential to improve diagnosis and drug development for these diseases.
Synucleinopathies are neurodegenerative diseases such as Parkinson's disease and dementia with Lewy bodies. They are associated with the accumulation of aggregates of misfolded alpha-synuclein proteins in the brain and spinal cord.
"When a misfolded alpha-synuclein protein in a neuron encounters one of its properly folded counterparts, it causes it to become misfolded," says Catherine Beauchemin of the RIKEN Center for Interdisciplinary Theoretical and Mathematical Sciences. "The two misfolded alpha-synuclein proteins stick together and cause other normal alpha-synuclein to misfold and aggregate."
This newly formed sticky seed continues to form more and larger aggregates, or fibrils, which spread to neighboring neurons.
"This is similar to a virus infection spreading from cell to cell," says Beauchemin, whose usual focus is viruses. "But while it's easy to swab a patient's nose to measure the concentration of a virus, for example, swabbing a patient's brain is not an option."
Synucleinopathies are often diagnosed late or misdiagnosed due to overlapping symptoms with other neurodegenerative disorders and a lack of robust biomarkers.
Seed amplification assays have emerged as promising diagnostic tools for these diseases. They can detect aggregating proteins not only from brain tissue samples of deceased patients, but also from less invasive samples such as skin scrapings or deep nasal swabs that can be obtained from patients living with synucleopathies.
Seed amplification assays are often used to determine simply whether a patient has aggregating fibrils or not. But more sophisticated assays can measure the amount of aggregating fibrils.
These latter assays involve preparing serial dilutions of a patient's sample, with multiple replicate reaction wells at each dilution, to identify the dilution at which half of the wells show signs of aggregation. A sample has a higher concentration of aggregating seeds if a greater dilution is required to stop aggregation.
These aggregating seed assays share similarities with another assay used to count infectious viruses, for which Beauchemin and her RIKEN colleagues have developed an improved concentration estimator2.
"Drawing from our previous work with viruses, we improved the performance of this seed amplification assay by reducing the dilution factor, increasing replicate numbers per dilution, and choosing an optimal algorithm to estimate the seed concentration," explains Beauchemin.
The team's results demonstrated significant improvements in the assay's reproducibility and quantitative accuracy.
"Our study highlights how assay design can markedly improve the measurement of disease-related alpha-synuclein aggregates across a variety of clinically relevant samples," says Beauchemin. "This should enable more precise evaluation of seeding activity to support important clinical and research applications."

Catherine Beauchemin and her colleagues have developed a refined test for quantifying alpha-synuclein aggregates with prion-like activity could improve the diagnosis of synucleinopathies such as Parkinson's disease. © 2025 RIKEN






