An innovative project to re-purpose existing drugs for their potential as antibiotics has uncovered a highly promising candidate with a potent and unique way of killing drug resistant bacteria.
A research collaboration between the University of Leiden, the Netherlands and the John Innes Centre, UK screened a chemical library of 352 small molecules for antimicrobial activity. These molecules have found uses in human therapies for cancer but had not been tested for their ability to kill harmful bacteria like Escherichia coli.
The screen identified 12 compounds that inhibited bacterial growth. The most potent of these, a representative of a class of compounds called isoquinoline sulfinamides, was chemically optimised by researchers at the University of Leiden.
The enhanced compound, LEI-800, exhibited antimicrobial activity against E. coli and K. pneumoniae, two gram-negative bacteria that often cause hospital acquired infections.
To find out why the compounds were able to kill these harmful bacteria, researchers selected naturally occurring resistant strains. They discovered that all the resistant strains contained mutations in genes that encoded the bacterial enzyme DNA gyrase, indicating this was the compound's target.
Gyrase is a bacterial topoisomerase: an enzyme that breaks and rejoins DNA to remove knots and entanglements and therefore is essential for bacterial growth.
Gyrase does not occur in humans and as such is a target of an essential fluoroquinolone antibiotics. However, the effectiveness of fluoroquinolones is severely compromised by growing bacterial resistance.
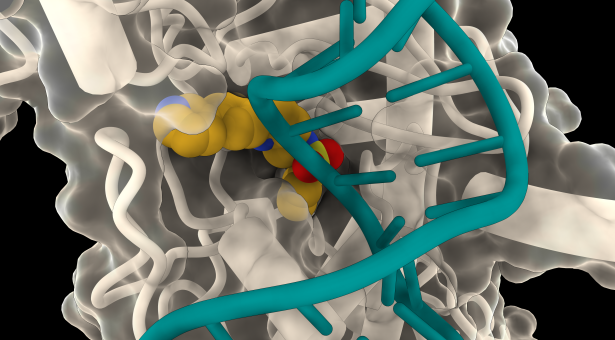
The John Innes team used cryo-EM microscopy to confirm that LEI-800 was indeed binding to E. coli gyrase and inhibiting it in an unprecedented way by preventing the enzyme from cutting DNA.
The search for new antibiotics is a global health emergency with new antibiotics urgently needed to keep pace with drug-resistant gram-negative bacteria. This study paves the way for the development of an entirely novel class of molecules targeting gram negative bacteria that have become resistant to existing drugs.
"It's exciting that we can find an entirely new type of antibiotic in this way," says one of the authors of the study Dr Dmitry Ghilarov from the John Innes Centre.
"Repurposing an existing library of successful compounds derived for other uses is a valuable approach because it takes away all the costs associated with a synthesis of a new library.
The unique binding pocket of the compound (the way in which it interacts with the target enzyme) means that the compound could offer an extra line of attack if used in tandem with existing antibiotics which are partially effective.
The approach outlined in this study promises more discoveries in the future, says Dr Ghilarov. "It is amazing to think that we screened a small library designed to target one class of enzymes, human kinases, and found an entirely novel topoisomerase inhibitor.
In terms of the chemistry available to drug discovery companies that is a small amount – they screen millions of compounds so the potential for drug discovery using this approach is enormous."
Well-known examples of drug repurposing include the anti-inflammatory drug aspirin that at low doses acts as a selective anti-blood clot drug, or sildenafil (Viagra) the erectile dysfunction drug that was originally developed to control blood pressure.
Dr Ghilarov said: "Biology is incredibly complex, there are thousands of different proteins in every cell presenting myriads of accessible surfaces, and it should not be surprising that a small molecule could bind to more than a single target.
"Repurposing is at the core of how humanity discovered medicines from nature: indeed, many anti-cancer drugs, such as doxorubicin or bleomycin are produced by soil bacteria Streptomyces to eliminate their bacterial competitors. "
Next steps for this research are to further optimise LEI-800 and move forward towards clinical trials and commercialisation.
Discovery of isoquinoline sulfonamides as allosteric gyrase inhibitors with activity against fluoroquinolone-resistant bacteria, appears in Nature Chemistry.
Image Caption – The figure shows the conserved horseshoe-shaped binding pocket on the surface of gyrase protein (white) and the bound LEI-800 (yellow spheres model). DNA is shown in transparent green.
Atomic model of this complex was determined by John Innes Centre scientists using cryogenic electron microscopy at Astbury Biostructure laboratory (University of Leeds)






