Whenever an organism develops and forms organs, a tumour creates metastases or the immune system becomes active in inflammation, cells migrate within the body. As they do, they interact with surrounding tissues which influence their function. The migrating cells react to biochemical signals, as well as to biophysical properties of their environment, for example whether a tissue is soft or stiff. Gaining detailed knowledge about such processes provides scientists with a basis for understanding medical conditions and developing treatment approaches.
A team of biologists and mathematicians at the Universities of Münster and Erlangen-Nürnberg has now developed a new method for analysing cell migration processes in living organisms. The researchers investigated how primordial germ cells whose mode of locomotion is similar to other migrating cell types, including cancer cells, behave in zebrafish embryos when deprived of their biochemical guidance cue. The team developed new software that makes it possible to merge three-dimensional microscopic images of multiple embryos in order to recognise patterns in the distribution of cells and thus highlight tissues that influence cell migration. With the help of the software, researchers determined domains that the cells either avoided, to which they responded by clustering, or in which they maintained their normal distribution. In this way, they identified a physical barrier at the border of the organism's future backbone where the cells changed their path. "We expect that our experimental approach and the newly developed tools will be of great benefit in research on developmental biology, cell biology and biomedicine," explains Prof Dr Erez Raz, a cell biologist and project director at the Center for Molecular Biology of Inflammation at Münster University. The study has been published in the journal "Science Advances".
Details on methods and results
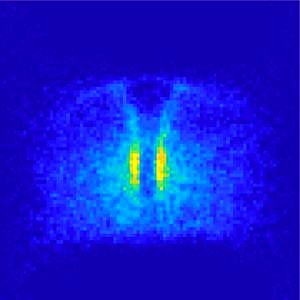
Researchers digitally merged microscopic images of hundreds of zebrafish embryos. This enabled them to detect patterns in primordial germ cell migration and to identify tissues that influence it. (Image: Gross-Thebing, Truszkowski, Tenbrinck et al. Sci Adv 2020;6: eabc5546/CC BY-NC)
For their investigations, the researchers made use of primordial germ cells in zebrafish embryos. Primordial germ cells are the precursors of sperm and egg cells and, during the development of many organisms, they migrate to the place where the reproductive organs form. Normally, these cells are guided by chemokines - i.e. attractants produced by surrounding cells that initiate signalling pathways by binding to receptors on the primordial germ cells. By genetically modifying the cells, the scientists deactivated the chemokine receptor Cxcr4b so that the cells remained motile but no longer migrated in a directional manner. "Our idea was that the distribution of the cells within the organism - when not being controlled by guidance cues - can provide clues as to which tissues influence cell migration, and then we can analyse the properties of these tissues," explains Łukasz Truszkowski, one of the three lead authors of the study.
"To obtain statistically significant data on the spatial distribution of the migrating cells, we needed to study several hundred zebrafish embryos, because at the developmental stage at which the cells are actively migrating, a single embryo has only around 20 primordial germ cells," says Sargon Groß-Thebing, also a first author and, like his colleague, a PhD student in the graduate programme of the Cells in Motion Interfaculty Centre at the University of Münster. In order to digitally merge the three-dimensional data of multiple embryos, the biology researchers joined forces with a team led by the mathematician Prof Dr Martin Burger, who was also conducting research at the University of Münster at that time and is now continuing the collaboration from the University of Erlangen-Nürnberg. The team developed a new software tool that pools the data automatically and recognises patterns in the distribution of primordial germ cells. The challenge was to account for the varying sizes and shapes of the individual zebrafish embryos and their precise three-dimensional orientation in the microscope images.
The software named "Landscape" aligns the images captured from all the embryos with each other. "Based on a segmentation of the cell nuclei, we can estimate the shape of the embryos and correct for their size. Afterwards, we adjust the orientation of the organisms", says mathematician Dr Daniel Tenbrinck, the third lead author of the study. In doing so, a tissue in the midline of the embryos serves as a reference structure which is marked by a tissue-specific expression of the so-called green fluorescent protein (GFP). In technical jargon the whole process is called image registration. The scientists verified the reliability of their algorithms by capturing several images of the same embryo, manipulating them with respect to size and image orientation, and testing the ability of the software to correct for the manipulations. To evaluate the ability of the software to recognise cell-accumulation patterns, they used microscopic images of normally developing embryos, in which the migrating cells accumulate at a known specific location in the embryo. The researchers also demonstrated that the software can be applied to embryos of another experimental model, embryos of the fruit fly Drosophila, which have a shape that is different from that of zebrafish embryos.
Using the new method, the researchers analysed the distribution of 21,000 primordial germ cells in 900 zebrafish embryos. As expected, the cells lacking a chemokine receptor were distributed in a pattern that differs from that observed in normal embryos. However, the cells were distributed in a distinct pattern that could not be recognised by monitoring single embryos. For example, in the midline of the embryo, the cells were absent. The researchers investigated that region more closely and found it to function as a physical barrier for the cells. When the cells came in contact with this border, they changed the distribution of actin protein within them, which in turn led to a change of cell migration direction and movement away from the barrier. A deeper understanding of how cells respond to physical barriers may be relevant in metastatic cancer cells that invade neighbouring tissues and where this process may be disrupted.
Funding
The study received financial support from the German Research Foundation (DFG), especially within the framework of the "Cells in Motion" Cluster of Excellence (2012-2019) and the Collaborative Research Center 1348 "Dynamic cellular interfaces: formation and function" at the University of Münster. In addition, funding from the Cells in Motion Interfaculty Centre and the Faculty of Medicine at the University of Münster, the European Molecular Biology Laboratory, the German Federal Ministry of Education and Research (BMBF) and the European Union (MSCA-RISE „Nonlocal Methods for Arbitrary Data Sources", NoMADS) contributed to the implementation of the study.
Original publication
Gross-Thebing S, Truszkowski L, Tenbrinck D, Sánchez-Iranzo H, Camelo C, Westerich KJ, Singh A, Maier P, Prengel J, Lange P, Hüwel J, Gaede F, Sasse R, Vos BE, Betz T, Matis M, Prevedel R, Luschnig S, Diz-Muñoz A, Burger M, Raz E. Using migrating cells as probes to illuminate features in live embryonic tissues. Sci Adv 2020;6: eabc5546.
- Original publication in "Science Advances"
- Software „Landscape" [en]
- Research group Prof Dr Erez Raz at the University of Münster
- Research group Prof Dr Martin Burger at FAU
- Research focus on "Cell Dynamics and Imaging" at the University of Münster






