The capacity of bacteria to spread disease across the Plant Kingdom may be much more widespread than previously suspected, according to new analysis.
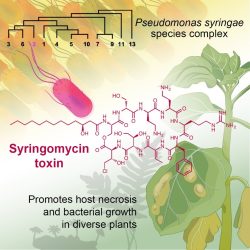
John Innes Centre researchers took a comparative evolutionary approach, using the diversity of Pseudomonas syringae bacteria, to determine how this pathogen infects distantly related plants.
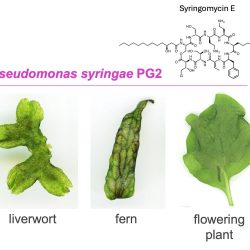
In experiments, researchers in the team of Dr Phil Carella, group leader, analysed the toxin syringomycin produced by the most widely infectious P. syringae strains, and compared its effect on both non-flowering and flowering plants.
The results showed that syringomycin was toxic in non-flowering plants (represented in this study by model species of a liverwort and fern), causing tissue death and activation of stress-related genes.
These effects were even more important for infections in non-flowering plants compared to flowering plants, which was surprising as much of our current understanding of how pathogenic (disease causing) bacteria manipulate plant hosts is centered on flowering plants which include some of our major crops.
By featuring non-flowering species, this study, which appears in Cell Host and Microbe, adds to a growing body of research that shows how bacterial pathogens carry the potential to colonise distantly related plants.
"Each of the plant species used in this study has a different life history since they last shared a common ancestor 500 million years ago. However, a single group of pathogens can infect each of them using a common set of pathogenicity factors," said Dr Carella.
"Our results demonstrate that pathogen virulence may be more general across plants than previously believed," he added.
The researchers hypothesise that P.syringae virulence is centered on fundamental processes shared amongst the Plant Kingdom. In this case, the toxin syringomycin likely interferes with cell membranes across each of the diverse plants tested.
Sometimes non-flowering plants are considered less sophisticated than their flowering relatives which arrived later in evolutionary history, but this study emphasises the importance of analysing the whole of the plant world to understand fundamental mechanisms and processes which could be applied to defending food crops against disease.
"Overall, our research shows that diverse plants can reveal useful knowledge about plant-pathogen interactions in general, which is informative for research on crop diseases. We don't eat liverworts, but they can teach us a lot about the core virulence mechanisms of important pathogens," observes Dr Carella.
The next step for this research is to explore the role the toxin plays in promoting the spread of bacteria, and how it cooperates with bacterial effector proteins to cause disease.
Another interesting research question to explore is why some P.syringae populations do not carry the toxin.
The group will also expand the diversity of plants used in the experiments to search for those that are resistant to bacterial pathogens.
A necrotizing toxin enables Pseudomonas syringae infection across evolutionarily divergent plants appears in Cell Host and Microbes.
Image Caption – Spores of Ceratopteris richardii, the model fern used in the study. Credit Phil Carella