
Salmonella has been at the centre of a scientific riddle that's puzzled researchers for more than 20 years.
The pathogen is well known for causing food-borne illnesses and can infect people through contaminated food or water. But until recently, no one knew how the microbe - strains of which can also cause typhoid fever and post-infectious arthritis - manipulates a fundamental class of cellular lipids called phosphoinositides.
Glenn Walpole, an MD/PhD student in the University of Toronto's Temerty Faculty of Medicine, is part of a team that solved the mystery. Their study was published in the journal Nature Cell Biology and featured on the cover of its May issue.
Walpole spoke with writer Erin Howe about the findings.
Tell us about your research and the question you set out to answer with this study.
During my PhD studies in the department of biochemistry, I explored how different microbes interact with human cells.
There are good microbes that help shape the development of our immune system, metabolism and several other important pathways. But my research focused on the small subset of microbes that are disease-causing, harmful to humans and which can be harmful to animals. This is the case with salmonella.
Researchers often use salmonella as a model system for intracellular pathogens, which can invade host cells and reside inside specialized compartments they create there.
These intracellular microbes secrete specialized proteins into human cells that manipulate our biology, which allows the pathogens to create a niche where they can extract nutrients, grow, replicate and even spread to other cells in our body. Several pathogens are capable of this type of behaviour, which is a challenge for our immune systems to recognize and clear.
Antibiotic-resistant forms of many pathogens that adapt to this intracellular lifestyle have recently been noted in the scientific literature; this adds to our concern, given that intracellular pathogens are a significant cause of death and disease around the world.
During my PhD, my supervisors and mentors (Sergio Grinstein, a senior scientist at The Hospital for Sick Children and professor in the department of biochemistry in the Temerty Faculty of Medicine; John Brumell co-director of the Inflammatory Bowel Disease Centre at SickKids and a professor of molecular genetics at U of T; and Greg Fairn, who is now in the department of pathology at Dalhousie University) and I wanted to study the initial stages of infection - when the pathogen invades the cells that line the gastrointestinal tract and set up residence.
We were interested in the lipids that form part of the membrane surrounding each and every cell in our bodies and how salmonella manipulates the lipids called phosphoinositides.
About 20 years ago, research showed that when salmonella infects a cell, it activates host signaling pathways that depend on phosphoinositide lipids. The host cell is 'tricked' into surviving despite having a harmful pathogen inside, which allows the bacteria to grow and replicate.
Similar survival pathways go awry in several types of cancer. It's almost as though the bacteria impose a cancer-like characteristic on the cell, by manipulating phosphoinositides. This is what we wanted to understand - because how salmonella was doing this had remained a real mystery to our field.
Two human epithelial cells [that] express a fluorescent biosensor specific for the lipid (in blue) were exposed to invasive salmonella (in red). In their study, Walpole and his collaborators reveal that this lipid, which supports bacterial entry and host survival, is generated by an unconventional phosphotransferase reaction driven by the secreted salmonella protein SopB.
Human epithelial cells expressing a fluorescent biosensor specific for the lipid (blue) were exposed to invasive salmonella (red).
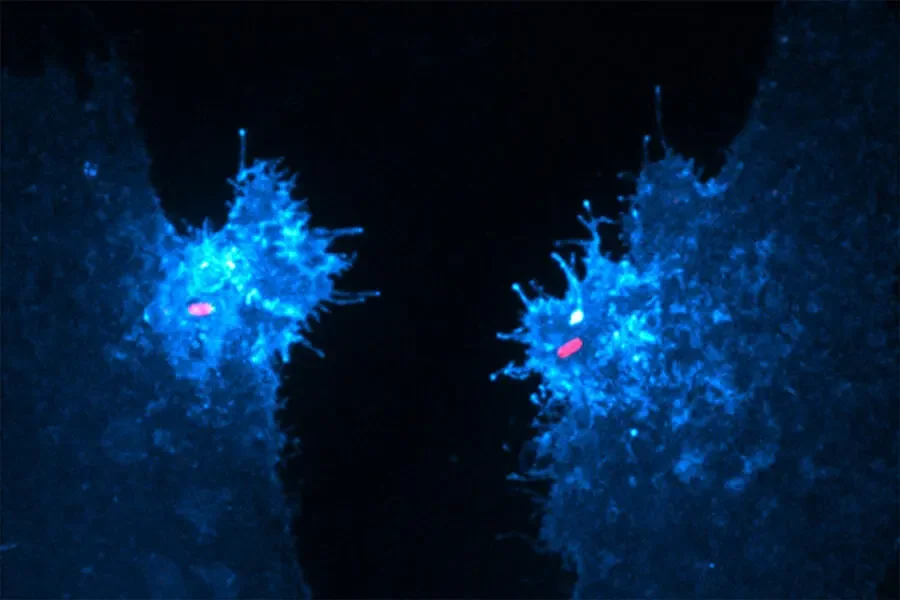
Human epithelial cells expressing a fluorescent biosensor specific for the lipid (blue) were exposed to invasive Salmonella (red).
So, what did you and the team discover?
We found that salmonella secretes a protein called SopB, which is like a little soldier that goes into the cell and manipulates phosphoinositides through a specialized enzymatic reaction. The reaction SopB performs on the lipid, called a phosphotransferase reaction, had never been described for any human or bacterial enzyme before this study.
This new knowledge is a breakthrough for the fields of host-pathogen interactions and cell biology. The implications are also quite interesting in terms of human biology.
What new pathways does this discovery open?
There are two new avenues we can explore from here. The first is the direct application to the infection process. By discovering a new enzymatic reaction, there's now a new target for treatment. Further research could help scientists discover or develop new antibiotics to target this specific enzymatic reaction.
And along those lines, we also discovered that a related protein secreted by a different pathogen - shigella, which also causes severe diarrhea - seems to have the ability to perform the same kind of reaction. So, we could target this reaction during salmonella infections, but potentially for other pathogens as well.
The second big possibility this study raises is whether the enzymatic reaction we discovered also takes place in human cells on an everyday basis in the absence of infection, and that we just never realized it.
Researchers who work in this field have carefully noted the primary pathways thought to be involved in forming these lipids such as PI3-kinases, another class of host enzymes. But our findings throw a curveball at our previous understanding. For example, several of these enzymes like PI3-Ks are drivers of certain forms of cancer. So, if human enzymes perform this additional phosphotransferase reaction, it could change the way that we think about this signaling pathway in cancer.
What has the response to your research been so far?
The response from the scientific community has been immensely positive. Through social media, for example, people are engaging with the paper and sending us questions and positive vibes from around the world, which is really cool. It's neat to see comments coming in from other researchers, either in the host-pathogen field, or who study phosphoinositides in the context of cancer or diabetes.
Also, when we uploaded this story in its preprint format, we received an invitation a day later to present at a worldwide zoom seminar series. I also presented this work at the Ontario Cell Biology Symposium, and we've been accepted to present in Lisbon, Portugal this coming summer. I think the field is excited about what our findings mean not only for host-pathogen interactions, but also for the many other processes that phosphoinositides regulate.
How exciting is it to make these kinds of waves when you are still completing the MD/PhD program?
It's very exciting, but it's important to remember an effort like this was really like being part of a football team. In addition to my supervisors, a really important collaborator and mentor of mine was our senior author, Fairn. He brought a lot of the technical expertise and creativity that we needed to probe such a difficult question.
And sometimes there's a bit of serendipity in science that isn't captured in publications. People have wondered about this question for 20 years since some of the initial work was published. It took perseverance to answer this question, but it also required the careful application of tools that didn't exist two decades ago.
We collaborated with a group at the University of Pittsburgh to do optogenetics with this enzyme, so we could activate the enzyme on demand with high energy light. This allowed us to see, on a minute-to-minute basis, what happened when we turned on the enzyme. This method only became available within the last few years.
We also did a type of lipid analysis called mass spectrometry with a group in England at the Babraham Institute. This approach to analyze the subset of phosphoinositides we were interested in was developed by that group and published only about two years ago. The findings in this study are truly the result of many people's efforts.
And what drew you to the MD/PhD program?
Pairing MD studies with doctorate research provides a really unique lens to view each field. Many fundamental insights into how people develop and experience illness and disease come from the clinic. Physician-scientists can take this forward and ask meaningful and important research questions from this lens. And here at Temerty Medicine, there are researchers in essentially every imaginable area of health research. The productivity is extraordinary.
What's next for you?
I'm finishing my second year in the MD program at the end of the month and looking forward to the sunshine this summer, travelling to Portugal and clerkship in a few months. I'll also be diving into a new area of research with Robert Inman and Nigil Haroon [both physicians in the University Health Network and faculty in rheumatology in the Temerty Faculty of Medicine] at the Schroeder Arthritis Institute in the coming months.






