The Drosophila Down syndrome cell adhesion molecule (Dscam1) can generate 38,016 different isoforms through alternative splicing, which has been illustrated as a classical example in many textbooks and treatises. It has been suggested as an important regulator of neural circuits. Each neuron acquires a unique cell surface identity through the expression of a unique combination of tens of isoforms in a largely stochastic manner, thereby mediating self-avoidance and the discrimination of self- versus non-self-neurites. These studies fail to elaborate on the remarkable bias of Dscam1. In contrast, highly biased splicing heavily circumscribes access to the full diversity from a pool of Dscam1 isoforms. The physiological significance of the highly specific bias in isoform expression remains elusive. More importantly, previous functional or phenotypic studies rely on genomic deletions or isoform overexpression. Therefore, it remains ambiguous whether these phenotypic defects observed in Dscam1 mutants are caused by changes in the number of isoforms, isoform bias, and/or protein levels.
The research team led by Prof. JIN Yongfeng from the Zhejiang University College of Life Sciences hit upon a brilliant idea about how to probe into the bias of Dscam1 while investigating its alternative splicing mechanism. The research is published in the journal Cell Reports.
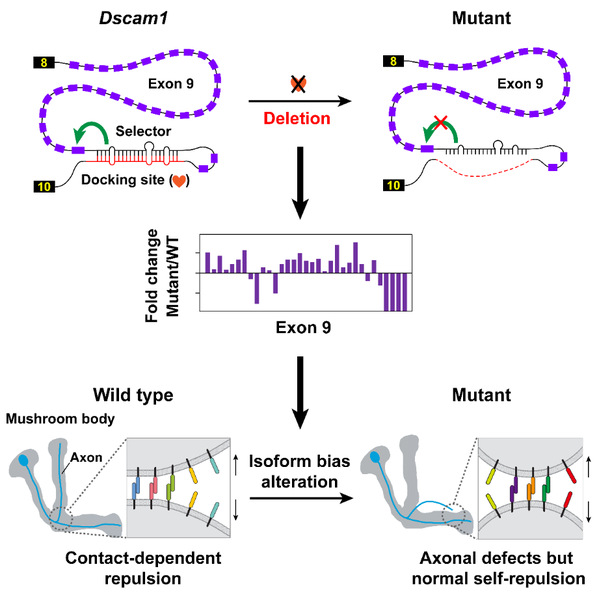
The study was initially aimed at examining the regulatory mechanism of Dscam1 alternative splicing. To this end, they deleted intronic cis-elements of Dscam1 alternative exons using the CRISPR-Cas9 system to analyze its effect on alternative splicing pattern. Evidence indicated that cis-elements mediated the regulation of alternative splicing by forming a competitive RNA secondary structure. This finding refuted the erroneous conclusion regarding Dscam1 alternative splicing recently published in Nucleic Acids Research. In that study, researchers claimed that Dscam1 alternative splicing was not regulated by the RNA secondary structure due to their failure to identify the RNA secondary structure.
Graphical abstract
Unexpectedly, Jin and his colleagues generated viable mutants with normal overall Dscam1protein levels and an identical number of Dscam1 isoforms, albeit showing a change in isoform bias. Phenotypic analyses revealed no significant variation in dendritic self-avoidance and self- versus non-self discrimination of Da neuron. Interestingly, mutants exhibited remarkable mushroom body defects, suggesting a variable domain-specific requirement for isoform bias. However, changes in isoform bias didn't influence the self-avoidance of axonal branches. The research team thus concluded that isoform bias may play a role in MB axonal wiring by influencing non-repulsive signaling in contrast to the isoform number that provides the molecular basis for neurite self-avoidance.
"Our research provides hereditary evidence for the elucidation of the role of Dscam1 alternative splicing bias in the neural system," said Jin. "More importantly, it suggests that Dscam1 isoforms differ from each other in function, opening the door to understanding the diversified neural functions of tens of thousands of Dscam1 alternative spliced isoforms."






