RICHLAND, Wash.-Quantum computers promise to speed calculations dramatically in some key areas such as computational chemistry and high-speed networking. But they're so different from today's computers that scientists need to figure out the best ways to feed them information to take full advantage. The data must be packed in new ways, customized for quantum treatment.
Researchers at the Department of Energy's Pacific Northwest National Laboratory have done just that, developing an algorithm specially designed to prepare data for a quantum system. The code, published recently on GitHub after being presented at the IEEE International Symposium on Parallel and Distributed Processing, cuts a key aspect of quantum prep work by 85 percent.
While the team demonstrated the technique previously, the latest research addresses a critical bottleneck related to scaling and shows that the approach is effective even on problems 50 times larger than possible with existing tools.
"Quantum computing can be extremely fast and efficient, but you have to address potential bottlenecks. Right now, preparing information for a quantum system is one factor holding us back," said Mahantesh Halappanavar, an author of the work and a member of the leadership team at the Center for AI @PNNL.
"This is a new way to package a problem so that a quantum computer can work on it efficiently," Halappanavar added.
The work is the result of a collaboration of several experts across the computing space at PNNL. Key contributors include S M Ferdous, an expert in high-performance computing and first author of the paper; Bo Peng, a quantum computing researcher; and Reece Neff, a graduate student at North Carolina State University and a distinguished graduate research program fellow at PNNL, who was the lead software developer for the project. Ferdous is a current Linus Pauling Distinguished Postdoctoral Fellow, and Peng is a former Pauling Fellow at PNNL.
Additional authors are Salman Shuvo and Sayak Mukherjee, with expertise in machine learning; Marco Minutoli, who specializes in high-performance computing; Karol Kowalski, laboratory fellow and expert in quantum computing; Michela Becchi of North Carolina State University; and Halappanavar.
To understand their work, imagine a restaurant with an award-winning menu and outstanding waiters. Without an efficient backroom kitchen operation that can keep up with the dishes and the cooking, it doesn't matter how good the food is if customers can't get a seat in the restaurant.
So it is with quantum computing: The backroom operations must run efficiently for quantum computers to achieve their potential.
"Quantum computing is not plug and play," said Peng. "You need to prepare the input a certain way so the quantum computer can understand and interact with it. Our algorithm is a tool for efficient hybrid computing, where we use classical computation to prepare quantum data for quantum computing."
Picasso slims the data
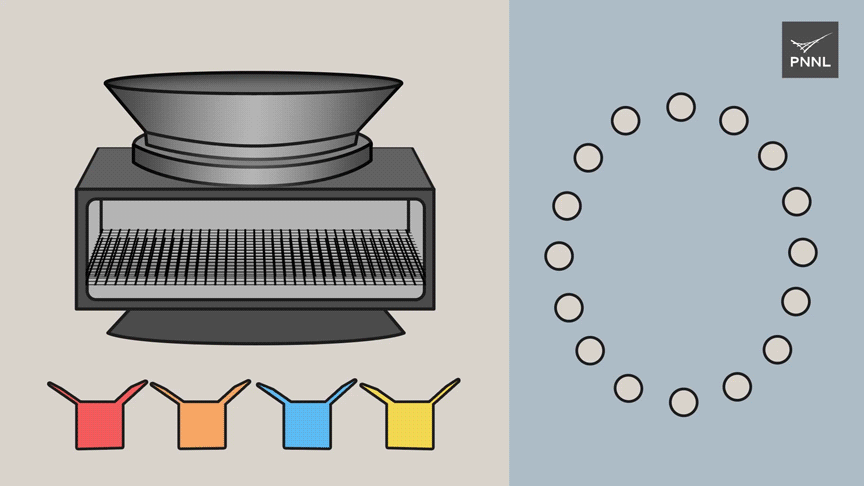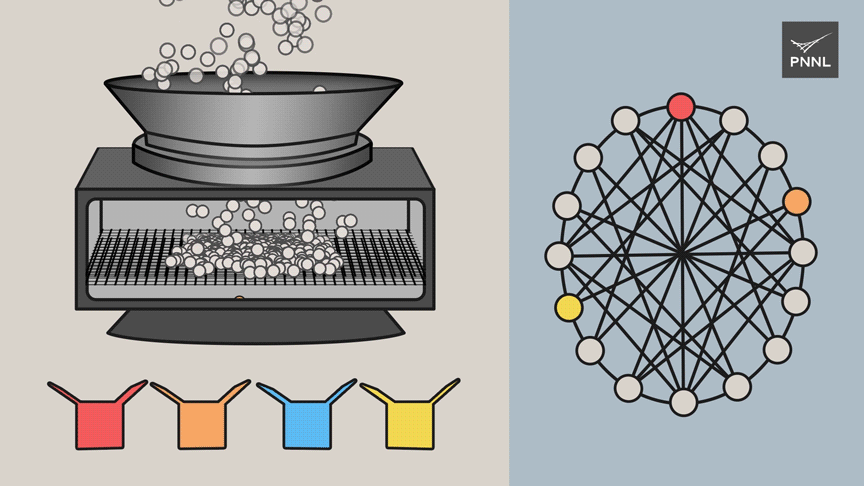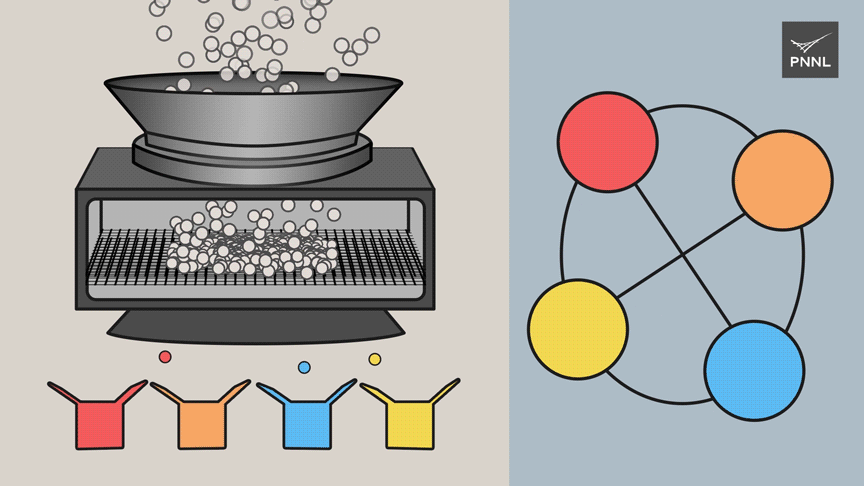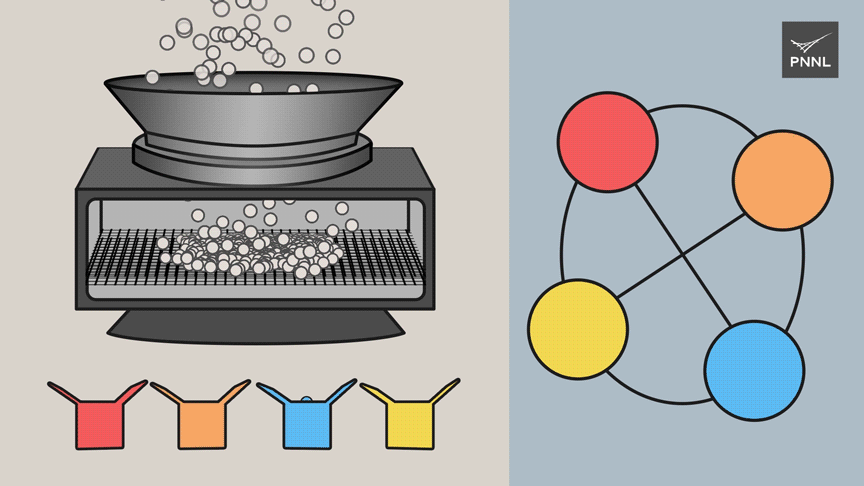
To lighten the computational burden, the PNNL team turned to a type of algorithm known as graph coloring-a specialty of Ferdous and Halappanavar. That approach allows researchers to explore relationships in a network and to quickly sort terms that are similar or different in some way. The goal is to sort all the relationships into as few groupings as possible.
The PNNL algorithm is named Picasso-a nod to the painter's use of color and the use of the term in graph analytics.
The team tested graph coloring in simulations of large hydrogen model systems, which are incredibly complex testbeds-simple chemical compositions that demand quick quantum data preparation requiring trillions upon trillions of calculations.
Some of the hydrogen systems the team tested generated more than 2 million quantum elements known as Pauli strings, translating to more than a trillion relationships for a classical computer to track. Current tools are typically limited to systems with tens of thousands of Pauli strings.
The PNNL team was able to lighten the computational load substantially by developing new graph analytics methods to group the Pauli operations, slashing the number of Pauli strings included in the calculation by about 85 percent. Altogether, the algorithm solved a problem with 2 million Pauli strings and a trillion-plus relationships in 15 minutes. Compared to other approaches, the team's algorithm can process input from nearly 50 times as many Pauli strings, or vertices, and more than 2,400 times as many relationships, or edges.
The scientists reduced the computational load through a technique known as clique partitioning. Instead of pulling along all the available data through each stage of computation, the team created a way to use a much smaller amount of the data to guide its calculations by sorting similar items into distinct groupings known as "cliques." The goal is to sort all data into the smallest number of cliques possible and still enable accurate calculations.
"From the perspective of high-performance computing, this type of problem really presents itself as a clique partitioning problem," said Ferdous. "We can represent an extremely large amount of data using graphic analytics and reduce the computation necessary."
Sparsification
The barrier to larger systems had been due to memory consumption, the researchers said.
"Pauli strings and their relationships quickly use up memory, limiting the size of the problems that can be tackled," said Ferdous. "Picasso uses modern tools such as streaming and randomization to sidestep the need to manipulate all the raw data."
The PNNL team's research work builds on work by other researchers who put forth a "Palette Sparsification Theorem" in 2019. Instead of including all the relationships between all the factors in their simulations, the PNNL team drew upon a much sparser dataset, only about one-tenth of the total, to perform accurate calculations. While the main data graph showed all the relationships among factors, the team's complementary, "sparser" graph showed only what the scientists call conflicts within the data. The conflicts alone supplied enough data for accurate calculations.
"You can set aside a great deal of data and still achieve an accurate result, using much less memory," said Ferdous.
Added Halappanavar: "It's like packing up a house for a move. You want to have the smallest number of boxes to move. You have to pack efficiently."
The researchers believe that Picasso can be extended to address even larger problems, including systems that require 100 to 1,000 qubits-the frontier of quantum computing.
As an added bonus, the team also developed an AI algorithm to help users calculate the best tradeoff between the amount of data used in the computations and the amount of memory required.
The work was funded by PNNL and by DOE's Office of Science.






