Kiwifruit flesh exhibits various colors, ranging from green to yellow, red and even purple. In kiwifruit, the red phenotype is the outcome of anthocyanin accumulation. Anthocyanin is one of the widespread pigments in nature and it plays crucial roles in plants and human beings. At present, several MYB transcription factors have been identified as having the potential of synthesizing anthocyanin in kiwifruit, but their specific regulatory functions and mechanisms have yet to be verified.

Recently, Prof. YIN Xueren and Dr. Wang Wenqiu at the Zhejiang University College of Agriculture and Biotechnology, in collaboration with Prof. Andrew C. Allan and his colleagues at the New Zealand Institute for Plant & Food Research Limited, and Prof. ZENG Lihui at Fujian Agriculture and Forestry University, published an article in New Phytologist, revealing key regulatory pathways for red-inner-pericarp and red-flesh kiwifruit by stable genetic transformation and molecular biology.
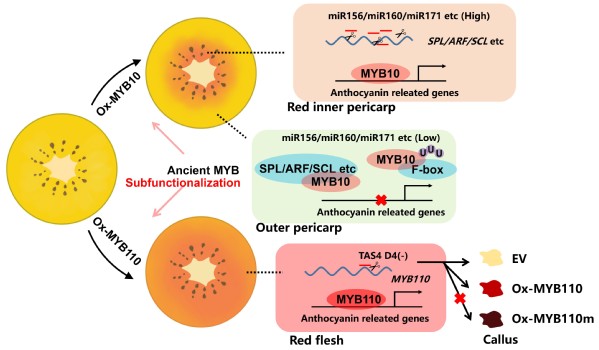
In this study, the researchers screened MYB10, a key regulator of anthocyanin synthesis in the kiwifruit with a red inner pericarp, by transcriptome analysis. Then they genetically transformed MYB10, as well as MYB110 from red-flesh kiwifruit, to obtain transgenic kiwifruit-the phenotypes of MYB10 (red inner pericarp) and MYB110 (red flesh). Whole-genome duplication (WGD) events suggested that MYB10 and MYB110 may originate from the same ancestor and develop sub-functions in the evolutionary history. Transient assays revealed that a motif at the N-terminal of MYB110 protein was essential for enhancing anthocyanin accumulation. Further experiments indicated that the transcripts of MYB110 were directly cleaved by a phasiRNA AcTAS4-D4(-), triggered by miR828. Although the MYB10 transcripts escaped this cleavage by AcTAS4-D4(-), the MYB10 protein was repressed by other regulators, which were the targets of miR156, miR160, miR171 and miR394. This investigation reveals the activation-repression systems for pigmentation in kiwifruit.






