A recent collaborative "PNAS" paper by the Allain (IBC), Jonas (IMBB), Leitner (IMSB) and Sharma groups (University of Arizona) reveals how early spliceosome complexes are brought together via specific RNA-binding of an unstructured arginine-glycine-glycine (RGG) motif.
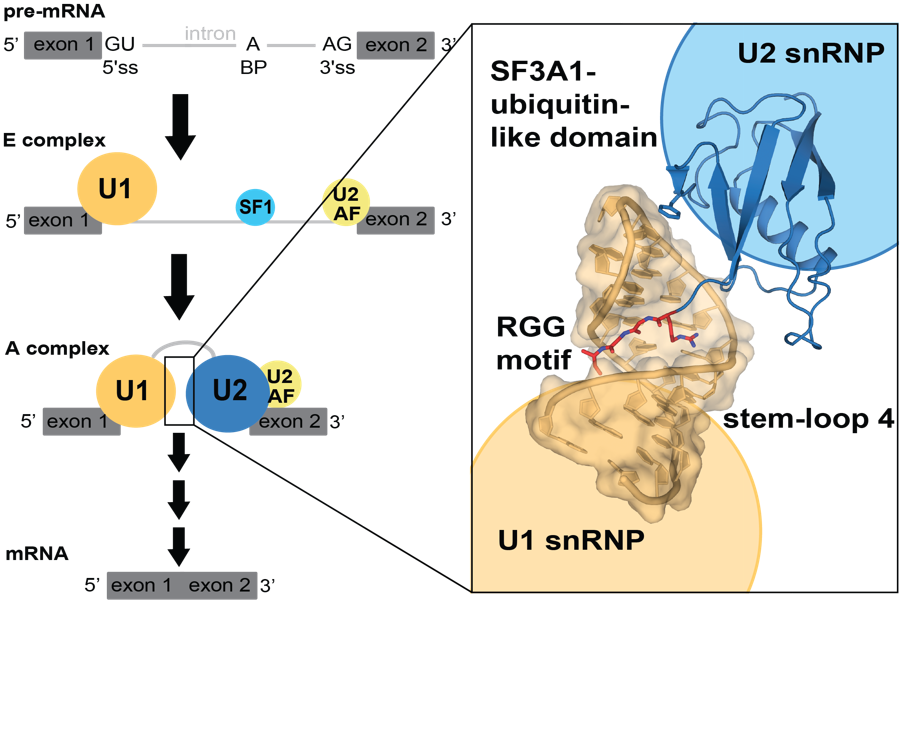
Pre-messenger RNA (pre-mRNA) splicing is a key regulatory step in gene expression. Malfunctioning of splicing causes several neurological diseases, such as spinal muscular atrophy (SMA) or amyotrophic lateral sclerosis (ALS). The splicing reaction is mediated by the spliceosome, a dynamic complex comprising five small nuclear ribonucleoproteins (snRNPs), which assembles onto each intron in multiple steps. During the early steps of spliceosome assembly, pre-mRNA bound U2 snRNP interacts with U1 snRNP for functional positioning of critical splicing elements. Thus far, this step has not been structurally characterized in mammals, even though it is a crucial step for splicing regulation.
In this study, the authors determined the 3D structure of the ubiquitin-like RNA-binding domain of U2 snRNP protein SF3A1 in complex with a stem-loop RNA of U1 snRNP. Crystallography, NMR spectroscopy, and cross-linking mass spectrometry revealed that the ubiquitin-like domain of SF3A1 recognizes sequence-specifically the RNA target. An RGG-motif of SF3A1 helps reading the RNA stem sequence via the arginine side chain and the main chain of both glycines.
The study is the first structural characterization of an ubiquitin-like domain and broadens the functional scope of unstructured RGG/RG-rich motifs in RNA binding proteins. Furthermore, the study provides a molecular basis of an early step of spliceosome assembly, which may help develop innovative therapeutic strategies against diseases originating from splicing defects.
Link to the paper in PNAScall_made.






