A step in the biochemical pathway for soapy plant compounds comes from the "wrong" set of tools

Soap-like plant compounds called saponins are used in the manufacture of drugs and vaccines, and they are exploited in the food industry, for example, to produce a low-calorie sweetener that's a thousand times sweeter than table sugar. Researchers at the Weizmann Institute of Science have revealed the entire chain of biochemical reactions by which saponins are synthesized in plants - and discovered that it includes a surprising case of molecular "hijacking." The scientists then performed a feat of synthetic biology, reproducing this chain by genetic engineering to cause a model plant and yeast to produce saponins. These findings open the way toward biochemical synthesis of potent saponins for use in industry and agriculture, for example, for producing targeted pharmaceutical additives, natural cosmetics and biological insecticides.
Saponins probably serve to protect plants from bacteria, fungi and insects; for one, they can kill insects by dissolving their lipid membranes. Historically, people have derived saponins from such plants as quinoa for use as soap or shampoo. In modern industries, too, saponins are chemically extracted from plants - for instance, from horse chestnuts to manufacture an anti-inflammatory ointment, from the soapbark tree Quillaja saponaria to make a vaccine booster and from licorice roots to produce the sweetener. But until now there's been no synthetic way of manufacturing saponins because it was unknown how they are made in nature.

Postdoctoral fellow Dr. Adam Jozwiak and other members of a team headed by Prof. Asaph Aharoni of the Plant and Environmental Sciences Department set out to learn how saponins are produced in spinach, which they found to be rich in these substances. The scientists separated out different types of spinach saponins and applied mass spectrometry to measure their mass and make predictions about their structure. They then purified some of the saponins to further analyze their structure using nuclear magnetic resonance. In parallel, they determined the expression patterns of the genes that take part in the synthesis of saponins.
These studies showed that creating saponins in spinach proceeds in ten major steps, each facilitated by its own enzyme. The scientists named these enzymes SOAP, numbering them from one to ten - SOAP1, SOAP2 and so on - and soon identified nine of them, but one enzyme, SOAP5, remained elusive. Its job is to attach a sugar tag called glucuronic acid at a specific spot in the saponin molecule. The problem was that attachment of sugar tags to such small molecules as saponins is known to be performed by a particular family of enzymes, yet no enzyme from this family performed this task in spinach.
Aharoni suggested that the mysterious SOAP5 might be an entirely different type of enzyme and pointed to one "suspect." It belonged to a family of enzymes that take part in making cellulose - not at all in the attachment of sugar tags to saponins - and are normally present within plant cell walls, not at all in the endoplasmic reticulum, the organelle in which saponins are produced.
The hunch proved spot on. The unlikely candidate, an enzyme from the cellulose synthesis family, indeed turned out to be SOAP5. The scientists confirmed this finding in several other plants that make large amounts of saponins, including soy, alfalfa, licorice, beetroot and quinoa.
The scientists have already engineered the entire set of saponin-making genes into a tobacco-like plant
"It was a major surprise to discover that an enzyme belonging to a 'wrong' family is involved in making saponins - just as you'd be surprised that, say, a carpenter is hammering in nails using a pipe wrench," says Jozwiak.
Aharoni suggests that this apparent misuse is a case of molecular "usurping": "This is an example of how, in the course of plant evolution, an essential enzyme was 'hijacked' to contribute to an unrelated function - production of substances involved in plant defenses."
Says Jozwiak: "Our findings may for the first time make it possible to synthesize saponins on a large scale in a sustainable manner, instead of depleting natural resources by extracting them from plants." In fact, the Weizmann scientists have already managed to engineer the entire set of ten saponin-making genes into a tobacco-like plant Nicotiana benthamiana, causing it to make one of the spinach saponins. Moreover, they managed to produce in yeast a smaller saponin molecule, the one used as the super-sweet sweetener.
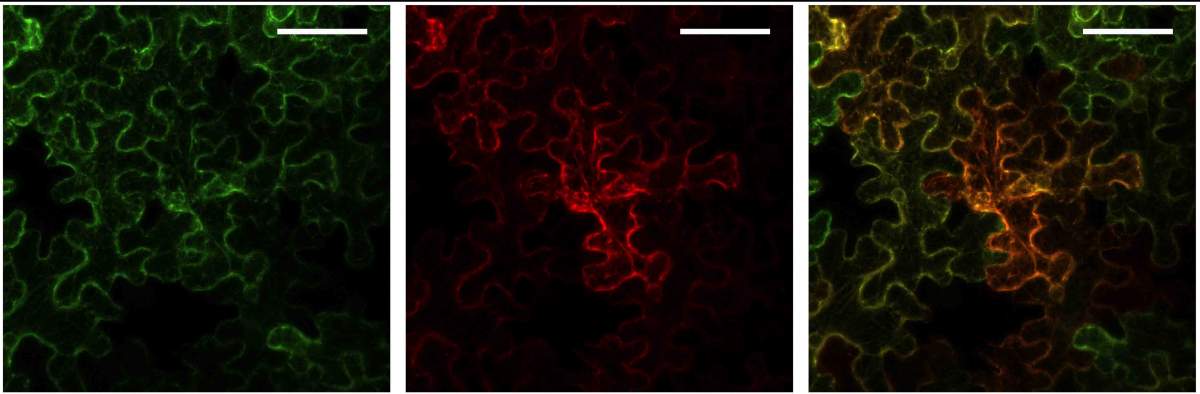
Conversely, the study's findings may help rid crops of the bitter taste of saponins by genetic engineering instead of having to wash them off, as is currently done, for example, with quinoa. This can be achieved by genetically neutralizing a single enzyme, SOAP5, which was shown to be essential to saponin production.
The research team included Dr. Prashant D. Sonawane, Dr. Sayantan Panda and Efrat Almekias-Siegl from the Plant and Environmental Sciences Department; Dr. Hassan Massalha from the Molecular Cell Biology Department; Dr. Tali Scherf from the Chemical Research Support Department; Dr. Constantine Garagounis and Prof. Kalliope K. Papadopoulou from the University of Thessaly in Larissa, Greece; and Dr. Bekele Abebie of the Agricultural Research Organization - Volcani Center in Rishon LeZion, Israel.
Prof. Asaph Aharoni is Head of the Vera and John Schwartz Family Center for Metabolic Biology; his research is also supported by the Benoziyo Endowment Fund for the Advancement of Science; the Knell Family Center for Microbiology; the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics; the Mary and Tom Beck - Canadian Center for Alternative Energy Research; the André Deloro Prize; the Laura Gurwin Flug Family Fund; the Tom and Sondra Rykoff Bioinformatics Facility for Plant Sciences; Dana and Yossie Hollander; the Sklare Family Foundation Cannabis Research Fund; the Yotam Project; the estate of Helen Nichunsky; the estate of Emile Mimran; and the estate of Betty Weneser. Prof. Aharoni is the incumbent of the Peter J. Cohn Professorial Chair.






